Introduction
Melon (Cucumis melo L., 2n = 2x = 24) is one of the most important horticultural crops with a great economic importance, belongs to the family Cucurbitaceae. The worldwide annual production of melon is around 30 million tons. China is the largest producer of melon in the world with the annual production of around 10 million tons1. In India, melon is produced approximately 1-2 million tons annually, the states Uttar Pradesh, Andhra Pradesh, and Punjab are the leading melon producers in the country where melon varieties such as Hara Madhu, Durgapura Madhu, Pusa Sharbati, Arka Rajhans, Arka Jeet, Pusa Madhuras and Pusa Rasraj are majorly grown for commercial cultivation2. The edible fruits of melon are very delicious, and rich in nutrients and minerals. The fruits of melon cultivars majorly vary in size, shape, pulp color, taste, sugar content and peel texture, thus exhibiting a huge polymorphism. So that, there is a great scope for conducting the breeding programmes in order to develop melon hybrids with the desirable traits3.
Generally, the plant breeding is a simple process that consists of three stages: (i) Selection of best parents; (ii) Crossing and producing the progeny; (iii) Recovery of progeny that outperform the parents4. Traditional plant breeding methods rely on phenotypic selection of progeny with superior traits. Though the genetic diversity is positively associated with phenotypic variation, yet also depends on environmental factors. As a result of that, the superior hybrids developed by the traditional plant breeding approaches often failed to perform well in the different environmental conditions. Therefore, for the successful development of a superior hybrid in any crop, the breeding programme should be planned to consider molecular plant breeding approaches in addition to the traditional selection of progeny based on phenotypic characters. Molecular breeding methods evaluate the genotypic variation and can assist in selection of superior hybrids that are consistent in carrying the variation in their phenotype as well as genotype, thereby eliminate the possibilities of human bias and save the time required for crop improvement4.
The genome sequence of C. melo was published in 2012. This draft genome sequence covers 83.3% of the total predicted melon genome size of 450 Mb, thus became a great genetic resource for the melon molecular breeders5. Since then, numerous molecular studies (for example: genome-wide association analysis, gene mapping and transcriptome profiling) that aimed to explore the molecular markers, novel genes and their characterizations have been conducted by utilizing the published melon genome sequence. Typically, the genome-wide association analysis and gene mapping studies are carried out by the following procedure: (i) Extraction of the genomic DNA of an individual plant; (ii) Whole-genome resequencing (or) Bulked segregant analysis (BSA) sequencing using primers designed based on the known genome sequence to identify linked single nucleotide polymorphisms (SNPs) and polymorphic markers (QTLs); (iii) Construction of a genetic map; (iv) Functional annotation and sequence analysis of the candidate gene based on the known genome sequence; (v) Characterization of the novel gene and validation of molecular maker in a natural population1, 6-12. Whereas, the transcriptome analysis is performed by the following procedure: (a) Extraction of the total RNA of an individual plant; (b) cDNA libraries construction and resequencing; (c) Quantitative Reverse Transcription PCR (qRT-PCR) using the primers designed for candidate genes based on the known genome sequence; (d) Identification and annotation of differentially expressed genes13-17. Altogether, these molecular studies would provide valuable genetic resources that can be directly used in molecular breeding for melon crop improvement.
In the present review, here we discussed the very recent research achievements of the crop improvement of Cucumis melo L. that including the studies on improving the melon fruit yield and quality, stress resistance, plant adaptability to climate changes, identification and characterization of novel functional genes, molecular markers, as well as understanding the genomics of plant physiology. In addition, this review presents the future perspectives of melon crop improvement.
Fruit Yield Of Melon
Melon hybrids for yield and other important traits, such as rind and flesh thickness, total soluble solids (TSS), β-carotene content and firmness can be produced by combining the novel inbred lines. 45 F1 muskmelon hybrids have been produced by crossing ten genetically diverse inbred lines (including two genetic male sterile lines) in a half-diallel mating design and evaluated for their fruit yield, level of phytochemicals and Fusarium wilt resistance. For the number of fruits per vine and TSS, the inbred lines KP₄HM-15 and MM-916 are found to be best general combiners. The highest fruit yield ha-1 and TSS was observed in the cross-combinations of MS-1 x M-610 and Kajri x MM-904, respectively. The highest standard heterosis for fruit yield and TSS is found in the hybrids KP₄HM-15 x MM Sel-103 and KP₄HM-15 x MM-1831. The inbred lines MM Sel-103 and KP₄HM-15 are generated the hybrids with highest resistance to Fusarium wilt. In addition, SSR marker analysis of these hybrids is identified the existence of genetic diversity among the parental lines used in this study18 (Table 1).
In Uzebekistan, the melon hybrids suitable of greenhouse cultivation are developed recently by crossing between a melon variety showing signs of male sterility and a female form of melon variety. The F1 hybrids namely, Zarkhal, L-161 x L-179 and L-161 x L-179 produced the larger fruits and considered for further variety testing inspection and production tests in greenhouses19. The success of melon hybridization programme majorly depends on choosing the highly genetically divergent parents. It is possible to obtain higher heterosis when more diverse parents are used. Therefore, the prior knowledge on genetic diversity of parental lines is paramount to undertake a worthful hybridization programme Multivariate analysis followed by Mahalanobis D2 statistics technique has been widely applied to evaluate the genetic divergence20. Recently, 35 muskmelon genotypes have been tested in randomized block design (RBD) were evaluated for genetic divergence by this technique and grouped into six clusters based on TSS, seed yield, days to appearance of first staminate flower, average fruit weight and the most divergent clusters are recommended for possible exploitation for future melon hybridization programme21 (Table 1).
At the horticulture station of the University of Tehran, Iran, the diallel crosses have been conducted using C. melo cultivars from Iran (Khatouni, Garmak, Abadan) and Japan (Jafa and Japan) in the 5 x 5 pattern in a randomized complete block design (RCBD) and estimated the combining ability for yield and fruit quality traits. Their analysis of general and specific combining abilities (GCA and SCA) was indicated the role played by the both additive and dominant gene effects in controlling the analyzed traits. The parent C. melo cultivar ‘Jafa’ showed the highest GCA for yield per plant and thus recommended for the breeding programme aiming to improve yield. Whereas the hybrid ‘Japan x Abadan’ exhibited relative high yield among all the hybrids produced in this study22 (Table 1). In another study, Line x Tester mating design was employed to understand the nature of gene action of various quantitative traits as well as to identify the suitable parents for further breeding programme. 30 F1 melon hybrids were generated by crossing the ten lines and three testers and performed the analysis of combining ability. The obtained results showed that the lines KM-1, KM-2 and KM-10 were found to be the best general combiners for fruit yield per hectare and three F1 hybrids were found to be recorded the highest fruit yield (Table 1). The observation of non-additive gene action for the most of the traits tested in this study indicating the great chances for melon crop improvement by the heterosis breeding23.
In India, at the Vegetables Research Farm of Banaras Hindu University, 26 genotypes of muskmelon germplasm were grown in an RBD design and calculated the genotypic and phenotypic variance of coefficients (GCV and PCV) as well as genetic variance (GA) for the various fruit yield and quality traits. Data revealed the highest PCV and GCV values and GA for the fruit yield per plant as compared to all other analyzed traits, and thereby suggested that the tested germplasm accessions have the great potential to be used in melon breeding programme as the parents24. Another study conducted at Anand Agricultural University, Gujarat, on the evaluation of 124 melon genotypes for their genetic variability found that the existence of high degree of variability among these genotypes as well as the highest GCV and PCV for the fruit yield per plant, TSS and fruit weight25. These 124 genotypes have also been evaluated for association analysis among 14 quantitative traits and observed that fruit length, weight, thickness showed significant and positive association with fruit yield per plant. Nonetheless, their path analysis carried out based on the genotypic correlation exhibited the positive and direct effect on various yield and yield contributing traits26.
Table 1: High fruit yield or quality melon cross combinations generated recently. (Numbers in the superscripts are corresponding to the cited references)
| High yielding hybrids of Melon | Characteristics |
| MS-1 x M-610 | High fruit yield18 |
| Kajri x MM-904 | Highest TSS18 |
| Zarkhal, L-161 x L-179 | Larger fruits19 |
| L-161 x L-179 | Larger fruits19 |
| Japan x Abadan | High fruit yield22 |
| KM2 x PS | High fruit yield23 |
In general, in some of the horticultural crops (for example: tomato, melon) the fruit shape and weight are closely related to the carpel number trait. The fruits with large diameter contain larger carpel number27. In melon, three-carpel number fruit is oval, whereas five-carpel number fruit is round in shape. However, the lack of information on the gene that regulate the carpel number trait was limiting the further research on melon fruit development. Therefore, the recent study of biparental genetic mapping of melon identified the candidate gene annotated as CmCLV3 is responsible for determining the carpel number. In three-carpel melon lines, the gene CmCLV3 was found to be expressed predominantly as compared to five-carpel melon lines (Fig. 1A). Thus, this study provides molecular breeding tools that target the carpel number trait for melon improvement6.
The plant growth regulator, Auxin regulates the fruit development by controlling the expression of the auxin early response gene family called SAUR (Small auxin up RNA). In melon, such SAUR family genes were recently identified by the genome-wide characterization. A total of 66 CmSAUR genes have been identified, which are observed to be highly homologous in the Cucurbitaceae plants. Further analysis of the transcriptome profiling revealed that CmSAUR19 and CmSAUR58 genes were found to be involved in fruit development, while CmSAUR38 and CmSAUR62 regulating the fruit ripening9 (Fig. 1B, 1C). The results of this study provided the insights into the future molecular breeding of melon targeting the early fruit development.
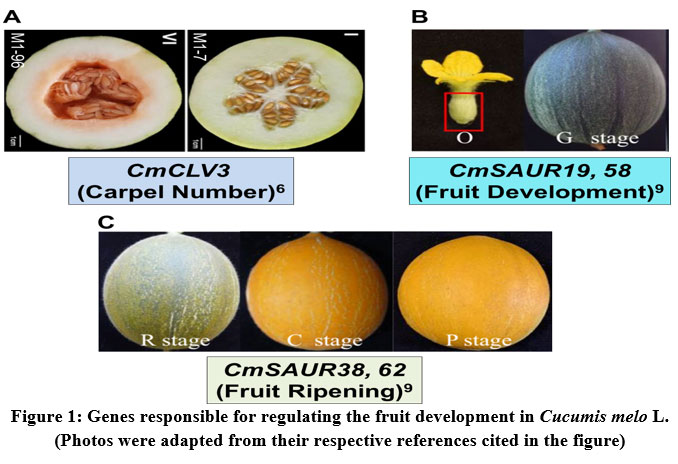 |
Figure 1: Genes responsible for regulating the fruit development in Cucumis melo L. (Photos were adapted from their respective references cited in the figure). |
Fruit Quality of Melon
Fruit acidity of melon affects its taste and volatility of aroma compounds, thus considered it as an important fruit quality trait. CmpH gene present on chromosome 8 determines the fruit acidity in melon and interestingly, its 12 bp duplication is associated with the loss of fruit acidity28 (Fig. 2A). Recently, a codominant molecular marker (DupI-12) has been designed at this duplication site and validated by performing an experimental analysis on screening of 92 Cucumis sp. accessions predominantly from India for their fruit acidity. The performed association analysis revealed the presence of significant association of this marker with fruit acidity as measured by titratable acidity3 (R2 = 73.43%) and pH (R2 = 70.76%).
Hami melon (C. melo var. saccharinus), a type of muskmelon majorly grown in the region of Hami, China, is a popular melon fruit for its excellent taste, sugar content and aroma. However, some of the hemi melon germplasms are less sweet with a different fruit shape. Hence, it is interesting to determine the genes that contributing to higher sugar content of the fruit. A recent comparative transcriptomic analysis of two hemi melons that differ significantly in fruit sugar content revealed the occurrence of a higher number of differentially expressed genes in the high fruit sugar content germplasm than the low fruit sugar content germplasm. Especially, the genes that involve in cell wall biogenesis and sugar metabolism (MUR4, UGD1) during fruit ripening were found to be highly expressed in the high fruit sugar content germplasm15 (Fig. 2B). The information from this study would help understand the underlying molecular mechanisms conferring higher fruit quality in melon and further development of fruit quality traits thorough the molecular breeding strategies.
Muskmelon seed coat color is one of the important horticultural traits, because it is related to flavonoids content and the antioxidant properties. Though the color of melon seed coat varies among different cultivars, yellow and brown colors are the most common ones. The brown seed coat contains relatively high amounts of flavonoids than the yellow seed coat. Nevertheless, the genetic basis for the seed coat color of melon remains unknown. Therefore, the recent study aimed at dissecting the genetic basis of seed coat color evaluated a six-generation population derived from the two parents with a yellow and a brown seed coat melon. The results from genome fine-mapping data revealed that a single gene CmBS-1 was found to be regulating the seed coat color. The gene expression of CmBS-1 was observed to be higher in brown seed coat than yellow seed coat melon, hence this gene can be exploited for the breeding programme of marker-assisted selection10 (Fig. 2C).
Fruit Morphology of Melon
Melon fruit firmness is one of the important horticultural traits that effects the storage, transport and fresh fruit consumption. A recent study on transcriptome profiling of the melon cultivar ‘Baogua’ provided new information on the melon genes associated with fruit firmness. The highest expression of CmGAL1 gene was observed at 21 days after pollination whereas CmGAL2–4 genes recorded highest expression at 14 days after pollination, as analyzed by the qRT-PCR analysis17 (Fig. 2D). This information is very useful for the melon breeding programmes aiming to improve the fruit firmness to keep maintain the fruit freshness while storing and transporting the fruits.
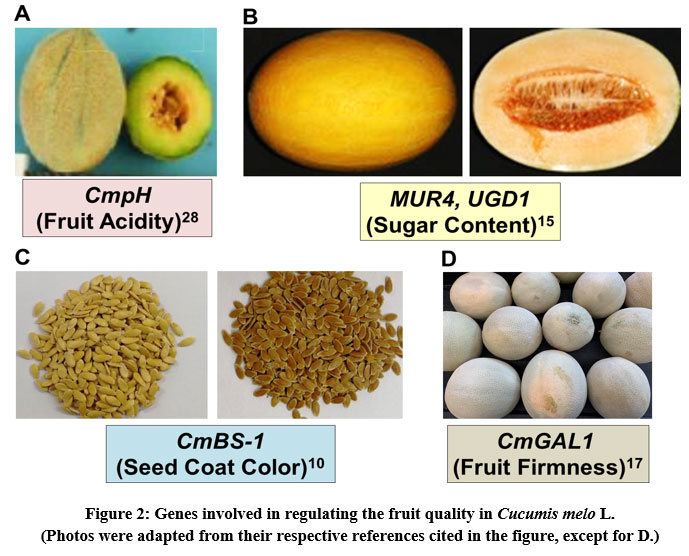 |
Figure 2: Genes involved in regulating the fruit quality in Cucumis melo L. (Photos were adapted from their respective references cited in the figure, except for D.) |
The length of fruit pedicel varies among the different melon cultivars. The melon fruit pedicel that connects the fruit to the plant body, determines the fruit appearance and quality. Hence, the understanding of the genetic basis for melon fruit pedicel length would be useful for future breeding programmes aimed at improving fruit quality. The recent whole genome resequencing of the melon identified a candidate gene on the chromosome 3, named CmFpl3.1. Its QTL was found to be linked to fruit pedicel length upon genotyping of 95 melon accessions that shows variations in fruit pedicel length1. Nonetheless, a recent comprehensive review on updates of melon breeding presents the list of QTLs identified for various morphological and biochemical traits of melon29.
Disease Resistance of Melon
The fungus Podosphaera xanthii is the causative agent of powdery mildew disease in melon. The disease symptoms include white power-like spots on leaves and fruits, and reduction in plant photosynthetic activity, which severely affect the plant growth and yields. However, some of the melon germplasm accessions have been reported to be resistant to powdery mildew. Therefore, the elucidation of the genetic basis for the powdery mildew resistance in the disease resistant accessions would provide valuable insights for the development of disease resistant melon varieties either by the hybridization or genetic engineering approaches. Recently, a study focused on transcriptome analysis of resistant melon cultivar TG-1 in comparison to the susceptible melon cultivar TG-5 revealed numerous candidate genes which might be conferring disease resistance either directly or indirectly. Especially, the transcription factors such as bHLH, ERF, MYB related and TALE were found to be differentially expressed in resistant cultivar (TG-1). Moreover, this study shed light on the potential role of the underlying signaling pathways such as PTI (PAMP-triggered immunity) and ABA (Abscisic acid) pathways that establishing the powdery mildew resistance in melon15. Another recent study was focused on developing the molecular marker required for the marker-assisted selection of melon hybrids resistant to powdery mildew. The molecular analysis of the resistant genotype (PMR 6) of melon for the race 5 of P. xanthii identified one major QTL named Pm-R. This Pm-R QTL can be used in the future disease resistance breeding programmes of melon to specifically select the hybrids with possible resistance to race 5 of P. xanthii30(Fig. 3).
The oomycete plant pathogen, Pseudoperonospora cubensis causes foliar chlorosis and necrosis in all members of the Cucurbitaceae family, hence this disease is named as cucurbit downy mildew (CDM). A recent study on QTL mapping for resistance to CDM identified a major QTL, qPcub-10.1, which can be used for future melon molecular breeding programmes that have goals to develop CDM resistant melon cultivars31.
A recent comprehensive review article on melon breeding for developing hybrids with resistance to powdery mildew and downy mildew has been discussed about the valuable melon germplasm accessions that can be used in resistance breeding32. The multiple resistance genes are found in various accessions of C. melo var. cantalupensis (PMR 45, PMR 5, PMR 6, and WMR 2) and C. melo var. momordica (PI 124111, PI 124112, and PI 414723), hence these accessions can be used as potential resistance donors in resistance breeding programmes32 (Fig. 3).
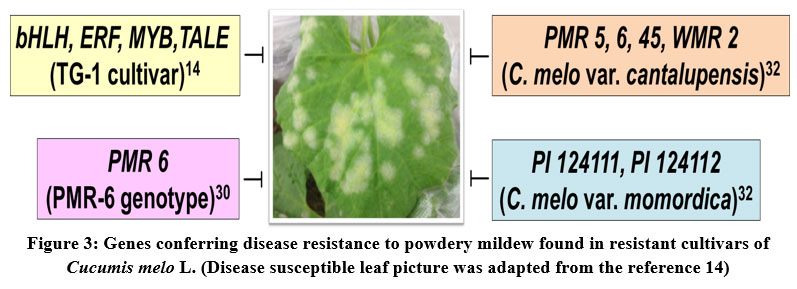 |
Figure 3: Genes conferring disease resistance to powdery mildew found in resistant cultivars of Cucumis melo L. (Disease susceptible leaf picture was adapted from the reference 14). |
Abiotic Stress Resistance of Melon
Plants growing in different environments and soil conditions would experience various abiotic stresses (salinity, osmotic stress, heavy metals etc.), but can able to tolerate such stresses to some extent with the help of their acclimatization mechanisms such as detoxification. The detoxification in plants is mediated by the family of proteins called multidrug and toxic compound extrusion (MATE) proteins. However, in melon, the information on such homologous MATE proteins was not available until now. The recent genome-wide characterization of MATE family in melon genome identified the 39 CmMATE proteins, thus provided the valuable information for the future functional genomics analysis to aim for the development of melon varieties tolerant to abiotic stresses7. Another similar study on MATE family of four Cucurbitaceae species (C. melo, C. sativus, Cucurbita pepo, and Lagenaria siceraria) identified 174 MATE genes and characterized their functions as transporters on plasma membrane, under severe salt stress8 (Table 2).
In addition to MATE proteins, there is another group of stress response proteins present in plants, the MYB transcription factors which play key roles in coffering plant resistance to autotoxicity and saline-alkali stress. Recent genome wide analysis studies identified and characterized the MYB genes in melon. A total of 178 MYBs were identified and they observed to be distributed in all 12 chromosomes of melon. Among these melon MYBs, 60 MYB genes are associated with autotoxicity whereas only 6 MYB genes are involved in saline alkali response. Interestingly, 2 MYB genes namely, CmMYB2R40 and CmMYB2R54 are responsive to above both types of stresses33 (Table 2). These results would provide key insights for breeding of abiotic stress-resistant melon varieties.
Table 2: Genes associated with abiotic stress resistance in Cucumis melo L. (Numbers in the superscripts are corresponding to the cited references).
| Type of Abiotic Stress | Genes involved |
| Salinity, osmotic stress, heavy metals7,8 | MATE |
| Low temperature11 | CmGLP2-5 |
| Autotoxicity33 | MYB |
| Saline alkali response33 | CmMYB2R40 |
Wider Adaptability of Melon
In order to develop the melon hybrids with greater resilience to climatic changes and to recommend the suitable genotypes for a particular region, the extensive research should be undertaken to evaluate the interactions between genotypes and environment (G x E). Recently, in Egypt, six new promising lines of C. melo were grown under six different environmental conditions considering (years x sowing dates) and evaluated the various traits. Interestingly, the data showed the existence of significant interactions between genotypes and environments, which indicated that genotypes behaved differently under different years. Among the melon lines tested, L-1 and L-4 genotypes showed wider adaptability to environment, and are recommended for cultivation in various environment conditions34.
Temperature stress in the plants affect their growth, photosynthesis and often leads to death. In order to cope up with the temperature stress, some of the plant proteins are specifically expressed in response to temperature stress. Germin-like protein (GLP) is one of the temperature responsive stress protein that regulate the plant stress due to low-temperatures. A genomic study focused on characterizing CmGLP gene family in melon has been found that CmGLP2-5 might be directly associated with low-temperature stress in melon11 (Table 2). These finding would be helpful in melon molecular breeding programmes for low-temperature stress tolerant cultivars.
In Indonesia, a new cultivar of melon named as ‘Meloni’ has been developed recently. In order to authenticate its new characteristics, the researchers followed the identification of molecular characters in addition to the traditional morphological characters. Because the morphological characters are highly plastic, mutagenic and varies with environmental conditions. But the molecular characters at the DNA level are stable and specific to that particular cultivar. The use of molecular markers, inter simple sequence repeats (ISSR) was carried out to detect the level of polymorphism and intra-species genetic variation in the ‘Meloni’ cultivar. These results provided the comprehensive information on the genetic stability and uniformity and confirmed its new characteristics at the molecular level35. Hence, this study suggests to follow both morphological and molecular identification of new characteristics in order to fulfil the requirements for the registration of a new melon cultivar.
Table 3: Melon species and their evaluated characteristics that discussed in this review. (Numbers in the superscripts are corresponding to the cited references).
| Melon species | Characteristics evaluated |
| C. melo var. chinensis | Seed coat colour10 |
| C. melo var. saccharinus | Fruit quality15 |
| C. melo ssp. agrestis | Fruit firmness17 |
| C. melo cv. Jafa | Fruit yield22 |
| C. melo var. cantalupensis | Disease resistance32 |
| C. melo var. momordica | Disease resistance32 |
| C. melo cv. Meloni | Genetic stability35 |
| C. melo var. makuwa | Transgenics43 |
Genomics of Melon Plant Physiology
C. melo is a monoecious plant, contains both male and female flowers present on the same plant. The sex determination in C. melo during the flower development from the primordia is regulated by the differential expression of sex determining genes (SDGs). The SDGs namely, CmWIP1 and CmACS11 promotes the male and female flower development, respectively36, 37. The proportion of male and female flowers is influenced by the position of the flower (female flower develops only at the most proximal nodes of lateral branches) at the optimal conditions but changes with variations in environmental factors38. At the molecular level, these changes drive the chromatin remodeling. The level of chromatin modification by histone methylation regulate the expression of a specific SDG, to selectively promote one sex type (either male or female) on the developing flower primordia. Specifically, the level of histone methylation of H3K27me3 is associated with the unisexual flower development in melon39. Recently, CmLHP1 proteins are found to be the key regulators responsible for determining the levels of H3K27me3, thus plays a major in sex determination in melon13.
Naturally, insect pollination by the honeybees (Apis mellifera, Apis cerana, Apis florea) takes place in the muskmelon, that helps in efficient fruit setting. The insect pollinators visit the plants when they are attracted by the flowers, and nectar. Stigma color of the flower is also one of the attracting features in the melon flower. Predominantly, there are two types of stigma colors such as green and yellow. Green stigma attracts the natural pollinators heavily than the yellow stigma. However, no information on genetic basis for the development of stigma color in melon is available. The recent comparative transcriptome analysis of melon lines with green and yellow stigmas provided the insights into the understanding of the regulation of stigma color at the gene level. Data from the transmission electron microscopy of chloroplast structures, RNA sequencing and transcriptome analysis, and qRT-PCR analysis, altogether concluded that relative expression of five key genes (CAO, CHLH, CRD, HEMA, POR) of chlorophyll biosynthesis pathway were observed to be highly expressed in green stigma melon than the yellow one16. The future breeding programmes aiming to increase the extent of natural pollination in melon by the insects could target the expression of these key genes. In addition, the recently identified QTLs linked to stigma color namely, SC2.1 and SC8.1 would further help in marker-assisted selection of a particular type of melon stigma color cultivars12.
Future Perspectives
The availability of melon genome sequence offers many opportunities for molecular breeders to research on the identification of novel genes, novel QTLs, and differentially expressed genes among the different populations. In addition, the screening of melon germplasm accessions for various fruit yield and quality traits, stress resistance, and wider adaptability to environments would help identification of novel germplasm resources that can be exploited for melon molecular breeding programmes. In case of melon traditional breeding, the existing major bottleneck is sexual incompatibility which is limiting the successful development of the inter-specific melon hybrids40, 41. To overcome this bottleneck, we should consider the molecular breeding approaches for melon such as marker-assisted selection and genetic engineering28, 42, 43. The genetic engineering by Agrobacterium tumefaciens mediated gene transfer, heterologous gene expression as well as the application of genome editing method called clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) would help develop the melon transgenic plants with the desirable traits44, 45. Moreover, the utilization of the latest speed breeding technologies would allow to study many generations within a short span of time and help develop the new varieties with desirable traits, quickly46.
Conclusion
Here, we have discussed the very recent achievements in the melon crop improvement, especially through the methods of molecular breeding approaches. In order to develop the new melon cultivars with the desirable traits, we must have to rely on the molecular methods in addition to the traditional breeding methods, together with these methods we can develop the stable melon cultivars within a short span of time. Selection of superior progeny in traditional breeding methods is only rely on the evaluation of phenotypic variation, which can be changed with changes in environment. Hence, the further selection of progeny should be carried out by evaluating its new characteristics at the molecular level, that would provide valuable insights into the stability of the genetic variation in the selected progeny. The current research on melon molecular breeding is focused on the identification of novel genes, novel QTLs, and their characterization and validation in order to expand the available melon genetic resources to exploit them in future marker-assisted selection as well as genetic engineering of melon. Nevertheless, the genetic dissection of melon germplasm resources for the desirable traits and the utilization of their natural polymorphism would also offer a great success in the future melon crop improvement.
Acknowledgement
Dr. M. Komala acknowledges Dr. Y.S.R. Horticultural University, Andhra Pradesh for providing all the infrastructure and research facilities. Dr. M. Komala would like to thank the Vice Chancellor of Dr. YSRHU – Dr. Tolety Janakiram, Associate Dean of COH, Chinalataripi – Dr. S. Sree Vijaya Padma and all of her colleagues for their support and encouragement. Mrs. Pragathi K would like to acknowledge her alma mater: College of Horticulture, Anantharajupet, Dr. YSRHU.
Conflict of Interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Funding Sources
There is no funding source.
References
- Cui, H., Ding, Z., Zhu, Z., Liu, S., Wang, X., Luan, F., Gao, P. Identification of major-effect QTL CmFpl3.1 controlling fruit pedicel length in melon (Cucumis melo ). Scientia Horticulturae, 2022; 110717.
CrossRef - Meena, N.K., Ram, L., Dangi, R., Choudhary, K., Prajapati, U. Scientific approaches in muskmelon cultivation: from field to market. CAU Farm Magazine, ISSN: 2279-0454, Silver Jubilee, 2018; 8(3):11–16.
- Dattha Reddy, G. P., Eguru, S. R., Lakshmanareddy, D. C., Sudhakar Rao, D. V., Yuvaraj, K. M., Syamsundar Reddy, P. Design and validation of a codominant molecular marker for fruit acidity in muskmelon (Cucumis melo). Plant Breeding, 2022; 2022:1–10.
CrossRef - Moose, S.P., Mumm, R.H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiology, 2008; 147:969–977.
CrossRef
- Garcia-Masa, J., Benjaka, A., Sanseverinoa, W., Bourgeoisa, M., Mira, G., Víctor, M., Gonzálezb., et al. The genome of melon (Cucumis melo) The Proceedings of the National Academy of Sciences, 2012; 109(29):11872–11877.
CrossRef - Wang, L., Wang, Y., Luan, F., Zhang, X., Zhao, J., Yang, Z., Liu, S. Biparental genetic mapping reveals that CmCLAVATA3 (CmCLV3) is responsible for the variation in carpel number in melon (Cucumis melo L.). Theoretical and Applied Genetics, 2022; 135:1909–1921.
CrossRef - Wang, S., Chen, K., Zhang, J., Wang, J., Li, H., Yang, X., Shi, Q. Genome-wide characterization of MATE family members in Cucumis melo and their expression profiles in response to abiotic and biotic stress. Horticultural Plant Journal, 2022; 8(4):474–488.
CrossRef - Shah, I.H., Manzoor, M.A., Sabir, I.A., Ashraf, M., Haq, F., Arif, S., Abdullah, M., Niu, Q., Zhang, Y. Genome-wide identification and comparative analysis of MATE gene family in Cucurbitaceae species and their regulatory role in melon (Cucumis melo) under salt stress Horticulture, Environment, and Biotechnology, 2022; 63:595–612.
CrossRef - Tian, Z., Han, J., Che, G., Hasi, A. Genome‑wide characterization and expression analysis of SAUR gene family in Melon (Cucumis melo) Planta, 2022; 255:123.
CrossRef - Hu, Z., Shi, X., Chen, X., Zheng, J., Zhang, A., Wang, H., Fu, Q. Fine‑mapping and identification of a candidate gene controlling seed coat color in melon (Cucumis melo var. chinensis Pangalo) Theoretical and Applied Genetics, 2022; 135:803–815.
CrossRef - Zhang, Z., Wen, Y., Yuan, L., Zhang, Y., Liu, J., Zhou, F., Wang, Q., Hu, X. Genome-wide identification, characterization, and expression analysis related to low-temperature stress of the CmGLP gene family in Cucumis melo International Journal of Molecular Sciences, 2022; 23:8190.
CrossRef - Lv, Y., Gao, P., Liu, S., Fang, X., Zhang, T., Liu, T., Amanullah, S., Wang, X., Luan, F. Genetic mapping and QTL analysis of stigma color in melon (Cucumis melo) Frontiers in Plant Science, 2022; 13:865082.
CrossRef - Granados, N.Y.R., Prado, J.S.R., Chaouche, R.B., An, J., Mianza, D.M., Sircar, S., Troadec, C. et al. CmLHP1 proteins play a key role in plant development and sex determination in melon (Cucumis melo). The Plant Journal, 2022; 109:1213–1228.
CrossRef - Zhao, Z., Dong, Y., Wang, J., Zhang, G., Zhang, Z., Zhang, A. et al. Comparative transcriptome analysis of melon (Cucumis melo) reveals candidate genes and pathways involved in powdery mildew resistance. Scientific Reports, 2022; 12:4936.
CrossRef - Liang, R., Su, Y., Qin, X., Gao, Z., Fu, Z., Qiu, H., Lin, X., Zhu, J. Comparative transcriptomic analysis of two Cucumis melo saccharinus germplasms differing in fruit physical and chemical characteristics. BMC Plant Biology, 2022; 22:193.
CrossRef - Lv, Y., Amanullah, S., Liu, S., Zhang, C., Liu, H., Zhu, Z. et al. Comparative transcriptome analysis identified key pathways and genes regulating differentiated stigma color in melon (Cucumis melo). International Journal of Molecular Sciences, 2022; 23:6721.
CrossRef - Zhang, H., Zhang, Y., Wang, P., Zhang, J. Transcriptome profiling of genes associated with fruit firmness in the melon variety ‘Baogua’ (Cucumis melo agrestis Jeffrey). Physiology and Molecular Biology of Plants, 2022; 28(2):301–313.
CrossRef - Kaur, S., Sharma, S.P., Sarao, N.K., Deol, J.K., Gill, R., Abd-Elsalam, K.A., Alghuthaymi, M.A., Hassan, M.M., Chawla, N. Heterosis and combining ability for fruit yield, sweetness, β-carotene, ascorbic acid, firmness and fusarium wilt resistance in muskmelon (Cucumis melo) involving genetic male sterile lines. Horticulturae, 2022; 8:2311–7524.
CrossRef - Ekaterina, L. Creation of heterosis hybrids of Cucumis melo for the protected soil of Uzbekistan. Eurasian Research Bulletin, 2022; 11:9–14.
- Mahalonobis, P.C. On the generalized distance in statistics. In: Proc. Nat. Acad. Sci. India, 1936; 2:49–55.
- Reddy, B.P.K., Begum, H., Sunil, N., Reddy, M.T. Genetic divergence analysis in muskmelon (Cucumis melo) International Journal of Current Microbiology and Applied Sciences, 2017; 6:2251–2260.
CrossRef - Esmaeili, M., Soltani, F., Bihamta, M.R., Nikkhah, M.J. Estimation of yield combining ability and fruit-related traits using diallel analysis in melon (Cucumis melo). International Journal of Horticultural Science and Technology, 2022; 9(1):131–142.
- Duradundi, S.K., Gasti, V.D., Mulge, R., Kerutagi, M.G., Masuthi, D.K.A. Combining ability studies in muskmelon (Cucumis melo) for quantitative and qualitative traits. The Pharma Innovation Journal, 2022; 11(7):2888–2893.
- Harish, K., Pal, A.K. Analysis of variance components for quantitative traits in muskmelon (Cucumis melo). Annals of Plant and Soil Research, 2022; 24(2):288–293.
- Prajapati, P.J., Acharya, R.R., Patel, N.D., Pandya, M.M. Studies on genetic variability, heritability and genetic advance in muskmelon (Cucumis melo ) genotypes. The Pharma Innovation Journal, 2022; 11(3):1683–1686.
- Prajapati, P.J., Acharya, R.R., Patel, N.D., Pandya, M.M., Patel, N.A. Character association and path analysis for fruit yield and it’s contributing traits in muskmelon (Cucumis melo). The Pharma Innovation Journal, 2022; 11(1):670–674.
CrossRef - Chu, Y.H., Jang, J.C., Huang, Z., van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct, 2019; 3:1–20.
CrossRef - Cohen, S., Itkin, M., Yeselson, Y., Tzuri, G., Portnoy, V., Harel-Baja, R., Lev, S. et al. The PH gene determines fruit acidity and contributes to the evolution of sweet melons. Nature Communications, 2014; 5:4026.
CrossRef - Kesh, H., Kaushik, P. Advances in melon (Cucumis melo) breeding: An update. Scientia Horticulturae, 2021; 282:110045.
CrossRef - Unlu, A., Polat, I., Yildirim, A., Onus, A.N. Mapping quantitative trait loci and developing first molecular marker for race 5 of Podosphera xanthii resistance in melon (Cucumis melo). Turkish Journal of Botany, 2022; 46:123–133.
CrossRef - Toporek, S.M., Branham, S.E., Katawczik, M.L., Keinath, A.P., Wechter, W.P. QTL mapping of resistance to Pseudoperonospora cubensis clade 1, mating type A2, in Cucumis melo. Theoretical and Applied Genetics, 2021; 134:2577–2586.
CrossRef - Cui, L., Siskos, L., Wang, C., Schouten, H.J., Visser, R.G.F., Bai, Y. Breeding melon (Cucumis melo) with resistance to powdery mildew and downy mildew. Horticultural Plant Journal, 2022; 8:1–17.
CrossRef - Zhang, Y., Xie, Z., Wang, F., Zhong, C., Liu, Y., Li, Z., Wang-Pruski, G., Zhang, Z. Genome-wide identification and characteristics analysis of melon (Cucumis melo) MYB transcription factors and their responses to autotoxicity and saline-alkali stress. Tropical Plant Biology, 2022; 15:93–109.
CrossRef - El- Sayed, A. A. Stability analysis for new lines of melon (Cucumis melo L.). Egyptian Journal of Applied Science, 2022; 37(1-2):18–28.
CrossRef - Yusuf, A.F., Wibowo, W.A., Daryono, B.S. Genetic stability of melon (Cucumis melo cv. Meloni) based on inter simple sequence repeat and phenotypic characteristics. Biodiversitas, 2022; 23(6):3042–3049.
CrossRef - Boualem, A., Troadec, C., Camps, C., Lemhemdi, A., Morin, H., Sari, M.A. et al. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science, 2015; 350:688–691.
CrossRef - Martin, A., Troadec, C., Boualem, A., Rajab, M., Fernandez, R., Morin, H., Pitrat, M., Dogimont, C., Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature, 2009; 461:1135–1138.
CrossRef - Harkess, A., Leebens-Mack, J. A century of sex determination in flowering plants. Journal of Heredity, 2016; 108: 69–77.
CrossRef - Latrasse, D., Rodriguez-Granados, N.Y., Veluchamy, A., Mariappan, K.G., Bevilacqua, C., Crapart, N. et al. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics and Chromatin, 2017; 10:1–17.
CrossRef - Robinson, R.W., Decker-Walters, D. Curcurbits. CAB International, 1999; Walling ford, Oxon, U.K. Saez, C., Esteras.
- Sheshadri, V.S., More, T.A. Indian land races in Cucumis melo. Acta Horticulturae, 2002; 588,187–193.
CrossRef - Li, P., Pei, Y., Sang, X., Ling, Y., Yang, Z., He, G. Transgenic indica rice expressing a bitter melon (Momordica charantia) class I chitinase gene (McCHIT1) confers enhanced resistance to Magnaporthe grisea and Rhizoctonia solani. European Journal of Plant Pathology, 2009; 125:533–543.
CrossRef - Bezirganoglu, I., Hwang, S., Fang, T.J., Shaw, J. Transgenic lines of melon (Cucumis melo var. makuwa cv. ‘Silver Light’) expressing antifungal protein and chitinase genes exhibit enhanced resistance to fungal pathogens. Plant Cell, Tissue and Organ Culture, 2013; 112:227–237.
CrossRef - Modrzejewski, D. Evidence Synthesis on the Impact of Genome Editing on Plant Breeding. 2020; PhD Thesis. Georg-August-Universitat Gottingen.
- Wu, H.W., Yu, T.A., Raja, J.A.J., Christopher, S.J., Wang, S.L., Yeh, S.D. Double virus resistance of transgenic oriental melon conferred by untranslatable chimeric construct carrying partial coat protein genes of two viruses. Plant Disease, 2010; 94:1341–1347.
CrossRef - Watson , A., Ghosh , S., Williams, M.J., Cuddy, W.S., Simmonds, J. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nature Plants, 2018; 4:23–29.

