Introduction
One of the major abiotic stresses that adversely affects plant growth, productivity, and quality, is soil salinity. About 20% of irrigated lands are negatively affected by salinity. In India, it was estimated that there are 6.74 million hectare of salt-affected lands that cause high economic losses. Among the various environmental stresses, soil salinity is one of the most devastating; it generally occurs either due to natural or anthropogenic causes and involves the accumulation of dissolved salts in the soil. In salt-affected areas, the crops suffer from stunted growth, poor biomass, and low yield, which are mainly attributed to osmotic stress, excessive absorption of sodium ions, generation of reactive oxygen species (ROS), alteration of metabolic processes, reduction of cell division and expansion, membrane disorganization, and genotoxicity.1 In addition, salt stress makes plants more prone to pathogenic diseases.2
Due to ground water scarcity in study location (Karur district, Tamil Nadu), villages nearby to Tamil Nadu Newsprint and Papers Limited (TNPL), depends on the usage of paper mill treated effluent as water source. These leads to high level of deposition of salt in this area. Therefore, in the study area, traditional farming is impractical due to the continues use of industrial treated effluent water as irrigation source, results in reduced crop yields. The high cost for physical remediation of salt issue makes conventional agriculture as economically unviable for most farmers in the region.3,4 Here, the primary agricultural activities traditionally involved the cultivation of paddy, sugarcane, plantain, groundnut, millet, maize, and corn. However, the use of industrial effluent for irrigation has resulted in the growth of only coconut and cattle grasses. Consequently, farmers with saline lands are exploring alternative income avenues through the implementation of agroforestry systems.5,6
Several reports focus on the cultivation of trees for improving soil fertility, plant health under stressed conditions, and an alternative income source for farmers. However, an excess of salt in the soil restricts the development of new roots, which is important for the growth of transplanted seedlings and the healthy development of trees under stressed conditions. The levels of salinity differ from site to site, and the salt tolerance capacity also differs greatly among the plant species.7 Planting of trees on such soils without considering their level of tolerance to salinity results in heavy mortality.8,9
A tremendous amount of fundamental research has focused exclusively on characterizing an array of salt stress-related genes in plants with the aim of improving plant salt tolerance using genetic modification approaches. However, these approaches have limited success and are highly expensive.10 Rhizosphere microorganisms as bio-inoculants are used as an eco-friendly and viable alternative to improve salt tolerance in plants and to ameliorate the soil salinity problems. The salt tolerance mechanisms of plant growth-promoting microorganisms isolated from saline habitat and halophilic bacteria that directly increase agricultural crop yields in salt-affected lands,11,12 are well understood. Halophilic (salt-loving) organisms have diverse physiological adaptive mechanisms to endure high salt concentrations.
Plant-associated microbial community could be the key for understanding the adaptation of plants to their habitat; they include PGPF such as Fusarium, Gliocladium, Trichoderma, Talaromyces, Phytophthora, Pencillum, Phoma, and Rhizoctonia. These increase plant growth, by improving the innate immunity and the levels of various important secondary metabolites in plants.13 High soil salinity impacts the growth of several plant species, particularly that of glycophytes (salt-sensitive) compared to that of halophytes (salt-tolerant plants). Therefore, it is not economical to grow salt-sensitive plants on extremely salt-affected lands. The use of salt-tolerant PGP microorganisms enhances the growth of salt-sensitive plants in salt-stressed areas. The apparent results of these beneficial microorganisms may not be evident under all natural conditions because of their insufficient naturally-occurring population in the soil.14 Therefore, the external application of the most suitable beneficial microorganisms becomes imminent. The effective utilization of bio-inoculants or bio-fertilizers in tree seedlings provides economic benefits, improves the soil fertility, and maintains the natural soil ecosystem.
There are only a few reports on the isolation and application of halo-tolerant fungal species, especially those from salt-affected agroforestry plantations. This study aimed to investigate the diversity of beneficial fungal communities in salt-affected areas, screen for their ability to support plant growth under salinity conditions, and evaluate the production of various compounds essential for processes critical for plant growth such as phosphate solubilization, potassium solubilization, zinc solubilization, nitrogen fixation, siderophore production, HCN production, ACC deaminase production, phyto-hormone production, and EPS production, under in vitro conditions.
Materials and Methods
Sample collection
The research site in Karur district is located in Tamil Nadu, South India, lies between 11°08’00” North and 77°98’39” South. The study sites are in close proximity to TNPL in Karur, where treated effluent from paper mill is utilized for irrigation. The introduction of this treated effluent has resulted in the fluctuations of soil physicochemical parameters. In response to increased salinity, local farmers have transitioned to agroforestry plantations. To enhance the growth of plants cultivated in this area, we have devised a plan to isolate and bio formulate native halotolerant PGPF from the rhizosphere of tree plantations.5,6
To implement the work, we collected rhizosphere soil samples from five distinct agroforestry plantations near TNPL Karur area. It was collected quarterly (Nov-Jan, Feb-Apr, May-Jul, and Aug-Oct) during the year 2016−2017. The selected plantations are as follows: (1) Kagithapuram Casuarina equisetifolia plantation (KACA), (2) Kagithapuram Eucalyptus camaldulensis plantation (KAEU), (3) Masagoundapudur C. equisetifolia plantation (MACA), (4) Moolimanglam C. equisetifolia plantation (MOCA), and (5) Murthypalayam E. camaldulensis plantation (MUEU). Soil samples were carefully collected in clean ziplock polybags and promptly transported to the laboratory, where they were stored at 4o C for subsequent analysis.
Physico-chemical analysis of collected soil samples
The soil samples collected from different study locations were analysed for their physico-chemical parameters such as pH, electrical conductivity (EC), and exchangeable sodium percentage (ESP) with three replicates, at the soil and water testing laboratory, Institute of Forest Genetics and Tree Breeding (IFGTB), Coimbatore by following standard procedures, as described by Dandwate.15
Enumeration of the total fungal count in rhizosphere soil
Standard procedures were employed for serial dilution and plating to quantify the population density of fungi in the soil samples collected.16 Ten grams of soil sample was weighed and used for serial dilution (up to 103). Then 0.1 ml of aliquots were serially transferred to sterile, labelled petri plates containing potato dextrose agar (PDA) and rose bengal agar (RBA). The plates were incubated at room temperature for 48 h. Three replicates were performed to access the Colony Forming Unit (CFU) per gram of soil sample.
Effect of different concentrations of sodium chloride (NaCl) on fungal growth
Due to fluctuations in soil salinity throughout the study period, we outlined a design to assess the maximum salinity tolerances of fungal isolates from various study locations. Accordingly, we conducted in-vitro tests to evaluate their varying levels of salinity tolerance by incorporating different NaCl concentration viz., 0% (control), 5%, 10%, 15%, and 20% by following the method prescribed by Ji.17 The fungal disc of 6 mm was placed on the center of PDA medium containing different concentration of NaCl and incubated at 28ºC for 48 h with three replicates. The growth of the fungal mycelium was compared with the control plate (without NaCl), and the percentage of mycelium growth was calculated. The isolate showing the maximum salt tolerance and the fastest growth was selected for further studies.18
Screening for qualitative PGP traits
Identification of phosphate- (P), potassium- (K), and zinc (Zn)-solubilizing isolates
Phosphate, potassium, and zinc solubilization under different NaCl concentration (0%, 5%, 10%, 15%, and 20%) was evaluated using tri-calcium phosphate as the insoluble phosphate (Pikovaskaya medium), mica powder as the insoluble form of potassium (Alexandero agar medium), and modified Bunt and Rovira agar medium containing 0.1% of ZnCO3 as the insoluble zinc source. Spot inoculation of the isolates was done at the center of the respective medium and the plates were incubated at 28±2°C for 10 days. The zone of clearance formed by the colonies was measured after every 24 h for up to 10 days. Each test was performed with three replicates. The solubilizing efficiency of the selected culture was calculated using the following formula as described by Sharma.19

Identification of nitrogen-fixing isolates
Nitrogen fixation was tested by inoculating selected fungal isolates on plates of N-free agar medium incorporated with different concentration of NaCl (0% (control), 5%, 10%, 15%, and 20%) and incubating the plates for 7 days at 28°C with 3 replicates. The isolates growing after repeated inoculation in N-free medium were considered to be positive for nitrogen fixation.20
Identification of siderophore-producing isolates
Siderophore production was assessed on solid chrome azurol S blue agar (CAS) plates with different concentrations of NaCl, as described by Hofmann.21 All the glassware used in the test were soaked in 2 N HCl solution for 24 h to avoid iron contamination from the glassware. Fungal disks were spot inoculated on the CAS blue agar plate and incubated at 28°C for 48 h with three replicates. The formation of a yellow-orange halo around the colony was considered as positive for siderophore production.
Identification of hydrogen cyanide (HCN)-producing isolates
HCN production was detected, as described by Joseph.22 All selected fungal isolates were inoculated in PDA, supplemented with glycine (4.4 g/l) and different concentration of NaCl. Whatman No. 1 filter paper was soaked with 2% sodium carbonate in 0.5% picric acid solution and placed at the top of the plate. Plates were sealed with parafilm and incubated at 28ºC for 4−7 days. Following incubation, the colour change of the filter paper was noted: yellow to brown or orange colour indicates positive result. The experiment was conducted with three replicates.
ACC deaminase-producing isolates
The assessment of ACC deaminase activity, followed the methodology outlined by Penrose and Glick.23 The colonies were screened for ACC deaminase activity on the sterile minimal Dworkin and Foster (DF) salts media supplemented with 3 mM ACC instead of (NH4)2SO4 as sole nitrogen source. Fungal disks were spot inoculated on the center of the plates and were incubated at 28°C for 3 days, with daily monitoring of mycelium growth. Fungal mycelium grown on the plates were recognized as ACC deaminase producers, while those lacking observable growth were classified as non-ACC deaminase producers.
Quantitative screening of PGP traits in selected isolates
The fungal isolates showing maximum PGP properties under high NaCl concentration were shortlisted and used for testing for important quantitative PGP properties. All the experiments were conducted with three replicates.
Quantitative estimation of IAA
We screened selected isolates for their ability to produce the plant growth hormone-IAA under in vitro conditions, as described by Turbat 24 and Bano and Musarrat.25 The test tubes containing PD broth supplemented with L-tryptophan (0.2 %) and different concentration of NaCl (0%, 5%, 10%, 15%, and 20%) were inoculated with selected fungal isolates and incubated for 7−8 days at room temperature. Following the incubation, the culture broth was centrifuged at 10,000 rpm for 30 min, and the pellet was discarded; 1 ml of the supernatant was transferred to a clean test tube. To this, 2 ml of freshly prepared Salkowski’s reagent was added. The tubes were incubated in the dark for 30 min and checked for the development of pink colour. Optical density at 530 nm (OD530) was recorded, and a graph was plotted against the standard values of IAA.
EPS production
EPS production was screened, as described by Ruhmann.26 Selected isolates were inoculated into 10 ml basal medium with different concentrations of NaCl (0%, 5%, 10%, 15%, and 20%) and incubated at room temperature for 5−6 days; 10 ml of culture suspension was collected and centrifuged at 3000 rpm for 15 min. Thrice the volume of chilled acetone was added to the supernatant. EPS was separated from the mixture in the form of a slimy precipitate; it was collected on a pre-dried filter paper, and the precipitates were allowed to dry overnight at 50°C. The dried filter paper was reweighed after overnight drying.
Morphological, cultural, and molecular characterization of PGPF
The cultural characteristics were studied using PDA and RBA; the colonies were observed for their macroscopic appearance (colour, reverse colour, pigmentation, growth rate, spore type, and hyphal arrangement). All selected isolates were subjected to microscopic analysis (microstructure).27 Fungal cultures were further identified based on 18S rRNA gene sequencing.28
Bioinformatics protocol
The 18S rRNA gene sequence analysis was performed for selected PGPF. DNA isolation was performed by using the EXpure Microbial DNA isolation kit developed by Bogar Bio Bee stores Pvt Ltd, Coimbatore, Tamil Nadu. Sanger’s di-deoxy nucleotide sequencing method was employed for the gene sequencing. Subsequently, a similarity search for the generated sequence was executed using the National Centre of Biotechnology Information (NCBI) with the help of BLAST program. The resulting sequences were deposited into the GenBank nucleotide database, and accession numbers were acquired. Following the retrieval of blast results, a phylogenetic analysis of the query sequence was conducted, which was subsequently followed by multiple sequence alignment with closely related sequences. Then the construction of the phylogenetic tree was accomplished using the Neighbour-joining (NJ) method.
Statistical analysis
PGP traits were analysed using one-way ANOVA followed by Duncan Multiple Range Test (DMRT) using SPSS software. Statistical significance was set at P < 0.05.
Results
Physico chemical properties of the rhizosphere soil collected from different study sites
Data on the physico-chemical properties of rhizosphere soil samples collected from different study locations. All the parameters varied during the quarterly intervals in the study period. Very high salinity was observed during the Feb-Apr season in all the study sites. The pH was alkaline (8.3) in the MOCA rhizosphere soil during Feb-Apr, compared to that in the other sites, which had a neutral pH. The pH ranged from 7.3−7.7 during Nov-Jan; 7.8−8.3, during Feb-Apr; 7.0−7.6, during May-Jul; and 7.0−7.7, during Aug-Oct. The soil samples collected from five different study locations showed distinct variations in EC throughout the study seasons. The EC values were the highest in the MUEU rhizosphere soil (17.5 dS/m) during Feb-Apr. The EC ranged from 1.4−4.7 dS/m during Nov-Jan; 4.4−7.5 dS/m, during Feb-Apr; 1.2−12.4 dS/m, during May-Jul; and 1.0−8.4 dS/m, during Aug-Oct interval. An EC >4 ds/m indicated that the soil was affected by salinity. ESP was below 15% at all the study sites (3.1−11.1%) (Table1).
Table 1: Physico-chemical properties of rhizosphere soil
| Parameters | Seasons | Study locations | ||||
| KACA | KAEU | MACA | MOCA | MUEU | ||
| pH | Nov-Jan | 7.7 | 7.4 | 7.7 | 7.3 | 7.4 |
| Feb-Apr | 7.9 | 8 | 7.8 | 8.3 | 8 | |
| May-Jul | 7.1 | 7 | 7.6 | 7.1 | 7.6 | |
| Aug-Oct | 7.7 | 7.5 | 7.5 | 7.3 | 7 | |
|
EC (dS/m) |
Nov-Jan | 4.7 | 4 | 1.4 | 2.2 | 2.6 |
| Feb-Apr | 13.2 | 4.4 | 11.9 | 16.5 | 17.5 | |
| May-Jul | 7.1 | 1.25 | 11.6 | 6.1 | 12.4 | |
| Aug-Oct | 6.2 | 1.05 | 8.4 | 5.1 | 8.1 | |
| ESP (%) | Nov-Jan | 10.5 | 8.4 | 3.1 | 4.6 | 4.2 |
| Feb-Apr | 9.1 | 8.1 | 8 | 7.9 | 8.5 | |
| May-Jul | 7.2 | 7.8 | 9.1 | 10.2 | 8.6 | |
| Aug-Oct | 9.1 | 8.2 | 9.5 | 9.3 | 11.1 | |
Values are mean of three replicates
Foot note: KACA- Kagithapuram C. equisetifolia rhizosphere soil, KAEU- Kagithapuram E. camaldulensis rhizosphere soil, MACA- Masagoundanpudur C. equisetifolia rhizosphere soil, MOCA- Moolimangalam C. equisetifolia rhizosphere soil, MUEU- Murthypalayam E. camaldulensis rhizosphere soil, EC-Electrical conductivity, ESP- Exchangeable sodium percentage
Population density of fungi
To understand the existence of microbial species in natural ecosystems, their isolation and characterization is important. The population density of fungi varied in all seasons at the different study locations. In PDA, the maximum population was detected from the MACA rhizosphere soil (30.2 x 103 cfu/g) during the Nov-Jan season, followed by that in the soil from KAEU (22.0 x 103 cfu/g) during the Aug-Oct season; a very low population was detected in the KACA rhizosphere soil (4.7 x 103 cfu/g) during the Feb-Apr season. In RBA, the maximum population was detected from the MUEU rhizosphere soil (19.5 x 103 cfu/g) during May-Jul, followed by that in the KACA rhizosphere soil (16.2 x103 cfu/g) during Nov-Jan; very less population was observed in the KACA rhizosphere soil (2.7 x103 cfu/g) during the May-Jul season. Therefore, Nov-Jan and May- Jun were optimum for fungal growth. PDA was more suitable for fungal growth than RBA ((Table 2).
Table 2: Population density of fungal colonies isolated from rhizosphere soils
| Sl
.No |
Location | Quarters of sample collection | Fungi (CFU 103 g-1)* | |
| PDA | RBA | |||
| 1 | KACA | Nov-Jan | 19.75b | 16.25b |
| Feb-Apr | 4.750a | 3.50a | ||
| May-Jul | 8.000a | 2.75a | ||
| Aug-Oct | 16.75b | 4.75a | ||
| 2 | KAEU | Nov-Jan | 20.75b | 15.00c |
| Feb-Apr | 13.50a | 10.00b | ||
| May-Jul | 12.25a | 8.250ab | ||
| Aug-Oct | 22.00b | 7.500a | ||
|
3 |
MACA |
Nov-Jan | 30.25c | 6.250a |
| Feb-Apr | 19.75b | 10.25c | ||
| May-Jul | 15.75a | 9.500b | ||
| Aug-Oct | 19.20b | 9.250b | ||
|
4 |
MOCA |
Nov-Jan | 16.50c | 13.00d |
| Feb-Apr | 7.250a | 6.500a | ||
| May-Jul | 11.25b | 7.000a | ||
| Aug-Oct | 12.50b | 10.00b | ||
| 5 | MUEU | Nov-Jan | 17.75b | 14.25b |
| Feb-Apr | 11.50a | 6.750a | ||
| May-Jul | 15.00ab | 19.50c | ||
| Aug-Oct | 16.00b | 14.50b | ||
Means, in a column, followed by common letter (s) are not significantly different at p<0.05 level according to DMRT.
Foot note: KACA- Kagithapuram C. equisetifolia rhizosphere soil, KAEU- Kagithapuram E. camaldulensis rhizosphere soil, MACA- Masagoundanpudur C. equisetifolia rhizosphere soil, MOCA- Moolimangalam C. equisetifolia rhizosphere soil, MUEU- Murthypalayam E. camaldulensis rhizosphere soil
A total of 68 fungal colonies with different morphology were selected from various study sites, and the pure cultures were stored for further use.
Screening for the NaCl tolerance of the isolated rhizosphere fungal colonies
Seventeen fungal isolates from the KACA plantation showed good mycelium growth in control (without NaCl) and in the 5% NaCl supplemented medium; 16 isolates, at 10%; 5 isolates, at 15%; and one isolate, at 20% salt concentration. In the KAEU plantation, 17 fungal isolates showed good growth in the control and in the 5% NaCl concentration; 11 isolates, at 10%; 3 isolates, at 15%, and only one isolate, at 20% NaCl concentration. Among the species from the MACA plantation, 11 fungal isolates were screened for NaCl tolerance. All isolates showed maximum mycelium growth in the control, and at 5% and 10% salt concentration; three isolates, at 15% and two isolates, at 20% NaCl concentration. In case of MOCA plantation, 12 fungal isolates showed maximum mycelium growth in the control, and at 5% and 10% salt concentration. Among the 12 isolates, 6 were tolerant to 15% NaCl and one isolate was tolerant to 20% NaCl. In the samples from MUEU plantation, 10 fungal isolates displayed maximum growth in the control and at 5% salt concentration; 7 isolates, at 10%; 5 isolates, at 15%; and 2 isolates, at 20% NaCl concentration. Number of fungal isolates showing NaCl tolerance is presented in Figure1.
 |
Figure: 1: Total number of salt-tolerant fungal isolates from different agroforestry plantations |
Foot note: C-medium without NaCl (0%), 5%-NaCl, 10%- NaCl, 15%- NaCl, 20%-NaCl
KACA- Kagithapuram C. equisetifolia rhizosphere soil, KAEU- Kagithapuram E. camaldulensis rhizosphere soil, MACA- Masagoundanpudur C. equisetifolia rhizosphere soil, MOCA- Moolimangalam C. equisetifolia rhizosphere soil, MUEU- Murthypalayam E. camaldulensis rhizosphere soil
Plant growth-promoting traits
The data on qualitative P solubilization are presented in Figure 2. Among the 7 fungi, F2 and F62 showed maximum P solubilization, F2 had the highest value at 27.00 SE (Solubilization Efficiency) followed by F62 (12.00 SE). Two isolates (F2 and F62) show P solubilization in 20% NaCl amended Pikovskaya agar medium.
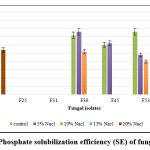 |
Figure 2: Phosphate solubilization efficiency (SE) of fungal isolates. |
The data on qualitative K solubilization are presented in Figure 3. The isolates, F2 and F36 displayed K solubilization at 20% salt concentration; F36 exhibited 40.00 SE followed by F2 with 12.00 SE. The isolates F2 and F36 show maximum K solubilization at 20% NaCl incorporated medium.
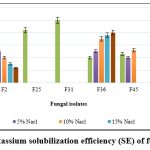 |
Figure 3: Potassium solubilization efficiency (SE) of fungal isolates |
The data on qualitative Zn solubilization are presented in Figure 4. Among the tested rhizosphere fungal isolates, none of the isolates showed Zn solubilization at 20% NaCl concentration. The isolate, F45 showed 15.00 SE in up to 15% NaCl; F25 shows 10.00 SE in 10% NaCl amended medium.
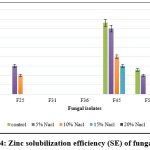 |
Figure 4: Zinc solubilization efficiency (SE) of fungal isolates |
The data on N fixation, siderophore, HCN, and ACC deaminase production are presented in Table 3. None of the fungal isolates in this study showed N fixation and ammonia production. Among those tested for HCN production, F45 and F53 produced HCN at the maximum NaCl 20% concentration. In case of qualitative production of siderophore, the fungal isolates, F2, F36, F45, F53, and F62 showed maximum production of siderophore up to 20% salt concentration. In case of qualitative production of ACC deaminase among the 7 fungal isolates, 5 (F2, F36, F45, F53, and F62) showed ACC deaminase activity at maximum concentration of NaCl. The isolates exhibiting the maximum qualitative traits were selected for further molecular identification.
 |
Table 3: Screening for qualitative plant growth-promoting traits of fungi |
Foot note: C- control (without NaCl), + positive result, – negative result
Quantitative production of IAA and EPS
F36 exhibited the maximum IAA production (50.6 μg/ml) at 20% NaCl concentration, followed by F2 (26.0 μg/ml), while F45 showed the least production (12.1 μg/ml) at maximum NaCl concentration; when the salt concentration was increased, the fungal isolates, F53 and F62 did not produce any IAA. EPS production was highest in F36 (3.54 g/ml) followed by F45 (2.1 g/ml) and F62 (1.56 g/ml) at 20% NaCl concentration. It was found that the increasing salt concentration affect the production of EPS in F53 isolate. The IAA and EPS production efficacy of selected isolates under different salt concentration is presented in Figures 5 and 6.
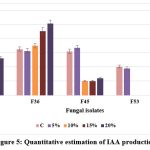 |
Figure 5: Quantitative estimation of IAA production |
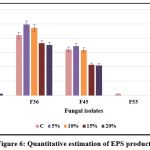 |
Figure 6: Quantitative estimation of EPS production |
Macroscopic and microscopic characterization of selected fungal isolates
Primarily, fungal colony morphology (macroscopic appearances) including mycelial colour, nature of pigments produced, growth rate, and microscopic characteristics including spore type and hyphal structure were evaluated (Table 4).
Table 4: Morphological and cultural characteristics of fungi
| Isolate code | Macroscopic appearance | Microscopic appearance | ||||
| Colony colour front view | Colony colour back view | Soluble pigment | Growth rate | Spore type | Hyphae | |
| F2 | Grey green colour velvety colonies | Light yellow | Absent | Slow | Conidia-globose to subglobose | Septate |
| F36 | Blue green colour, concentric rings present | Pale yellow | Absent | Rapid | Conidia-globose | Septate |
| F45 | Light green colour powdery colonies | Pale yellow | Absent | Rapid | Conidia-globose | Septate |
| F53 | White puffy colonies | Bluish green colour with concentric white ring | Absent | Slow | Conidia-Verrucose | Septate |
| F62 | White cottony colonies | White colour | Absent | Moderate | Ameroconidi-cylindrical | Septate |
Molecular characterization of selected PGP microbes
The PGP fungal isolates showing maximum salt tolerance and in vitro plant growth-promoting activity (F2, F36, F45, F53, and F62) were selected for molecular identification. The 18S rRNA genes were successfully amplified using PCR. The BLAST comparison searches with NCBI nucleotide database revealed 99% similarity of the isolate F2 with Pencillum citrinum (Genbank accession number: MF170859), F36 with Trichoderma viride F1 (Genbank accession number: MF170862), F45 with Trichoderma viride F2 (Genbank accession number: MF170858), F53 with Periconia variicolor (Genbank accession number: MF170861), and F62 with Acremonium borodinense (also known as Bulbithecium borodinense) (Genbank accession number: MF170860) (Table 5).
Table 5: List of salt-tolerant fungi identified using 18s rRNA sequencing
| Sl.
no |
Isolate code | Organism isolation site and sample | Organism identified as | Genbank accession number |
| 1 | F2 | Kagithapuram C. equsetifolia rhizosphere soil | Penicillium citrinum | MF170859 |
| 2 | F36 | Kagithapuram Eucalyptus rhizosphere soil | Trichoderma viride F1 | MF170862 |
| 3 | F45 | Masagoundanpudur Casuraina equsetifolia rhizosphere soil | Trichoderma viride F2 | MF170858 |
| 4 | F53 | Moolimangalam Casuraina equsetifolia rhizosphere soil | Periconia variicolor | MF170861 |
| 5 | F62 | Murthypalayam Eucalyptus rhizosphere soil | Acremonium borodinense (Bulbithecium borodinense) | MF170860 |
The phylogenetic tree of the PGP fungal isolates was constructed using neighbor-joining bootstrap analysis (Figure 7a,7b,7c,7d,7e). The identified colonies are shown in Figure 8.
 |
Figure 7a: Neighbor-joining tree for F3 isolate |
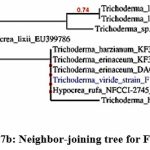 |
Figure 7b: Neighbor-joining tree for F36 isolate |
 |
Figure7c: Neighbor-joining tree for F45 isolate |
 |
Figure7d: Neighbor-joining tree for F53 isolate |
 |
Figure7e: Neighbor-joining tree for F62 isolate |
Foot Note: Neighbor-joining tree based on the 16S rRNA gene sequence showing the phylogenetic relationship of F2, F36, F45, F53, and F62 to related isolates
 |
Figure 8: Salt-tolerant plant growth promoting rhizosphere fungi |
Discussion
Physico-chemical parameters of soil samples
The physico-chemical properties of five different soil samples, collected from selected agroforestry plantation sites at quarterly intervals were evaluated. The pH of soil samples was neutral throughout the study time. The soil nutrient parameters were not constant across the different study locations during the study period; this could be attributed to differences in irrigation water, application of chemical fertilizers, temperature, and rainfall. Similar results were reported earlier for tropical soils.29 Rousk30 reported variations in soil pH, due to different biotic and abiotic factors during their study period.
The soil with EC >4 dS/m, ESP >15%, and pH <8.5 is considered as saline or salt affected soil (U.S salinity laboratory staff, 1954). In this study, EC > 4 dS/m was observed in most of the soil samples at the various study sites. Among the four seasons, the soil samples collected during summer (Feb-May) showed the highest level of EC. This may be due to less rainfall, saline water irrigation, especially the use of industrial treated effluents, use of chemical fertilizer, and high temperature. These promote the accumulation of soluble salts and their retention near the soil surface, resulting in higher EC. Similar results were reported by Iwai31 increased level of EC from 1.80 to 28.60 dS/m during the dry season. Sharma32 reported that the higher evaporation during dry seasons results in the accumulation of soluble salts at the upper surface of the soil owing to capillary rise. Kawser33 reported the variation in the salinity of soil samples due to climate change throughout the year; low salinity was observed during rainy season and high salinity was observed during the dry winter and summer seasons, in the upper soil horizons; these results are similar to that obtained in this study.
Diversity status of microorganisms in salt-affected soil
Several PGP microbes are used as bio-fertilizers worldwide, contributing to increasing soil fertility and plant growth and yield. Primary metabolites such as amino acids, carbohydrates, organic acids, glucosinolates, and vitamins, as well as secondary metabolites such as alkaloids, flavonoids, phenolics, sulfur-containing compounds, and terpenoids, are secreted from the roots of crop plants, and they help the rhizosphere microflora to develop, communicate, and interact with one another. The beneficial microbial communities are better attracted to and increased by the exudates, which also destroy pathogenic organisms by preventing their growth and invasion.34 It was found that, numerous soil factors impact the microbial community and its functions, thereby influencing nutrient availability and plant growth.
In this study, 68 rhizosphere fungal isolates were obtained from different rhizosphere regions. The population density of fungi varied among and within the different locations and periods. This could be attributed to the varying parameters such as physico-chemical properties of the soil, environmental conditions, salinity, and temperature. Msimbira and Smith35 reported that the biological activity and composition of soil microbes are influenced by various factors including physico-chemical properties of soil, temperature, and vegetation. Ponmurugan and Gopi36 described that the variation in population density of different beneficial microbes could be attributed to several reasons such as soil nutrients, pH levels, moisture content, organic matter, and certain soil enzyme activities. Similar to the results of this study, Grishkan37 stated that the hypersaline desert soils have a less richness of fungi. Similarly, Ibekwe38 suggested that changes in microbial density could be the first indicator of stress in saline or salt-affected soils. Zhang39 showed that salinity has a negative impact on microbial abundance, diversity, composition, and functions.
In this study, we aimed to determine the salt tolerance ability of soil fungi under in vitro conditions. Among the 68 soil fungi studied, only seven fungal isolates showed tolerance to 20% NaCl; a steady state of growth was observed in the maximum isolates up to 10% salt concentration. There was a drastic reduction of number of colonies observed when the salt concentration was increased above 10%. The reduction in population density with increasing concentration of NaCl could be attributed to hyper-osmolarity, which results in reduction of microbial cytoplasmic water activities. Dave and Desai40 isolated bacteria and fungi at different NaCl concentrations from marine salterns, Bhavnagar, Gujarat, India, and they reported similar results; the total microbial count and overall diversity of beneficial microbes decreased with increasing NaCl concentration. Oren,41 documented that halophiles can be categorized into three groups based on their salt tolerance levels: slight halophiles, moderate halophiles, and extreme halophiles. Slight halophiles exhibit optimal growth within a 0.2–0.5 M (1–3%) NaCl concentration. Moderate halophiles thrive in environments with a 0.5–2.5 M (3–15%) NaCl concentration, while extreme halophiles can endure and grow in conditions with a higher salt concentration, ranging from 2.5–5.2 M (15–30%) NaCl. In the present investigation, out of the 68 isolates examined, only 7 (10.29 % isolates) demonstrated a notable salt tolerance of up to 20% NaCl concentration. Interestingly, among these, 5 isolates exhibited multiple PGP activities in conditions of elevated salt concentration. Effectively utilizing this extreme halotolerant PGP isolates in saline-affected soil can significantly enhance plant growth and productivity.
Mayak42 suggested that the selection of beneficial microbial isolates from naturally-stressed environment or rhizosphere is a possible measure for improving crop health because it can be used to control diseases and promote plant growth. Choudhary43 suggested that stress can be detrimental for sensitive microorganisms; it reduces the activity of surviving cells, owing to the metabolic load imposed by stress tolerance mechanisms. Arit44 reported that the adverse effects of high salt concentrations in soil leads to dispersion and flocculation of soil particles; in addition, it affects organic matter solubility and its availability. This affects the microbial community structure. In a previous study,45 we isolated 16 salt-tolerant bacterial isolates from the rhizosphere soil of Casuarina equsetifolia and the isolates showed a maximum salt tolerance of up to 12% NaCl.
Screening for PGP traits
PGP microbes under salt stress exhibit positive effects on plants; they positively influence the germination percentage, drought tolerance, shoot and root weight, plant growth, and yield. Abbas46 described that the plants grown in salt-affected soil harbor diverse group of microbes in their rhizosphere, which have the potential to cope with salinity. These halotolerant microbes assist plants in withstanding the increased concentration of salt through the production of different organic and inorganic compounds such as IAA, ACC deaminase, and volatile compounds.
In this study, nitrogen fixation, and the production of siderophore, ammonia, ACC deminase, IAA, and EPS were analyzed to prove the PGP ability of selected fungal isolates. Plant growth is improved by the mineralization of nutrients, mediated by PGPF, in the soil. It reduces the breakdown of the substrates into soluble molecules that can be absorbed by the plants. An essential microelement that supports plant growth is phosphours (P). In this study, P solubilization was evaluated using Pikovskaya agar medium, in which a fungal isolate (F2) showed 27.00 SE (Solubilization efficacy) at 20% NaCl concentration. There was a wide variation in the solubilization of P under different salt concentrations. Srividya47 studied the effect of salinity on the phosphate solubilization by A. niger F7 isolated from agricultural soils, and the isolate showed different levels of P solubilization under different saline conditions; it could tolerate a maximum salinity of 2 % NaCl; this level of tolerance is low when compared to that exhibited in this study. Malgioglio48 reported that the complex organic phosphorous was solubilized and mineralized into different small molecules by PGPF, through the stimulation of enzymes like phosphatases, phytase, organic acids, and inorganic acids.
K solubilization was carried out in Aleksandrov agar medium. Among the different fungal isolates, F36 showed the maximum K solubilization efficacy (40.00) at 20% NaCl concentration, when compared with the others. Similarly, Bhattacharya49 isolated K solubilizing microbes from saline soil. Parmar and Sindhu50 reported that the soil beneficial microorganisms such as bacteria, fungi, and actinomycetes solubilize the insoluble potassium to soluble forms through different mechanisms such as acidolysis, chelation, exchange reactions, and the production of inorganic and organic acids and polysaccharides. Decomposition of organic matter in the soils leads to the production of formic acid, citric acid, and oxalic acid. These organic acids increase the solubilization of K compounds by providing protons and calcium ion complex.51
Zinc is a micronutrient essential for plants in very low amounts. In this study,
Zn solubilization was carried out by using Bunt and Rovira agar medium; no fungal isolates exhibited Zn solubilization at 5,10, 15 and 20% NaCl concentration. Tavallali52 reported that the deficiency of Zinc leads to susceptibility to heat, light, and fungal infections; in addition, it affects grain yield, pollen formation, root development, and water uptake in agricultural crops. Saravanan53 reported that the production of siderophore leads to Zn solubilization. Zn solubilizing microorganisms solubilize zinc through various mechanisms. These microbes produce anions by lowering the pH and chelate zinc for solubilization. Among the different PGPF, Trichoderma sp. promotes the absorption of minerals and nutrients, mainly, Fe, K, N, and P.54
HCN production enhances the availability of P and other elements for plant growth. Therefore, HCN production by the microbes at high salt concentration is considered as one of the significant plant growth promoting traits. We determined the HCN production by different fungal isolates. The HCN production facilitates antibacterial and antifungal activity on plant pathogens. Anand 55 reported that HCN could be toxic for plant pathogens. Rijavec 56 highlighted that HCN is a bio-control agent against fungal pathogens. It has the ability to form complexes with transitional metals in the mineral substrate;57 the ecological function of HCN is based on its interference with the release of elements from the mineral substrate. Recent studies on mineral weathering in natural environments58,59 indicated that HCN-producing bacteria (HPB) promoted the mobilization of elements from rocks forming the minerals. HCN production is reported in various PGPFs such as Aspergillus niger, Penicillium sp., and Trichoderma sp.60-63
In this study, the amount of siderophore production gradually increased with increase in salt concentration up to a certain level. All isolates showed siderophore production, which was a significant observation. Iron is the fourth major requirement for plants as well as microbes but is unavailable for direct assimilation. Ferric ion is pre-dominant in nature, and it is sparingly soluble (10−18 M); this makes its concentration too low to support plant growth. Therefore, plants and microbes sequester biologically available iron through secretion of siderophores in iron-stressed conditions. Crowley 64 reported siderophore production in two PGPF, P. chrysogenum and R. arrhizus; this upgraded Fe content and promoted growth in cucumber, maize, and tomato plants, and augmented the chlorophyll content through an increase in Fe-EDTA availability. The production of siderophores by a number of PGPF, including Absidia spp., Aspergillus spp., Thermoascus aurantiacus, Aspergillus flavus, A. niger, A. tamarii, A. nidulans, Fusarium sp., Paecilomyces varioti, Cunninghamella sp., and Penicillium are extensively reported.65,66
All selected PGP isolates were positive for ACC deaminase activity. Synthesis of ACC deaminase under different salinity ranges, 5 to 20% NaCl, indicates its role in plant growth by reducing ethylene stress. Plant growth promotion by selected PGP microbes, irrespective of salinity can be correlated with the higher production of the ACC deaminase enzyme. Similar results were reported by Zhang 67 and Viterbo 68 in Trichoderma spp., which are known for their potential to hydrolyse ACC into ammonia for plant growth. ACC deaminase is found in Trichoderma sp. The inoculation of plants with PGP microbes that express ACC deaminase activity facilitate extensive root growth in seedlings due to the reduction in ethylene stress;69 this imparts resistance to crop plants against various stresses including salinity.70
All PGPF were screened for IAA production under in vitro conditions. Among 5 isolates, F36 and F2 exhibited the highest IAA production at a 20% NaCl concentration, surpassing the others. In contrast, the remaining isolates demonstrated a notable decline in IAA production as the salt concentration increased. IAA, a plant hormone, is a natural auxin produced by PGPF. The microbes isolated from salt-affected soils can tolerate salt stress. Numerous investigations have illustrated the role of microorganisms in triggering indole-3-acetic acid (IAA) signaling pathways and augmenting the endogenous IAA levels in plants grown in salt stressed areas.71 Microbial phytohormones modulate physiological processes such as root cell growth and tissue differentiation and boost rhizosphere microbes.72,73 Murali60 isolated Penicillium janczewskiy and reported that it can inhibit Rhizoctonia solani, a plant pathogen that causes stem rot, through IAA production. Various fungal endophytes, including Aspergillus flavus, Aspergillus niger, Fusarium oxysporum, Penicillium corylophilum, Paecilomyces Formosus and Penicillium funiculosum have been documented for their ability to promote plant growth and produce indole-3-acetic acid (IAA) as reported by Deng and Cao.74 Additionally, Muhammad75 highlighted the capacity of Penicillium roqueforti to synthesize IAA, thereby assisting host plant species in coping with stressful saline conditions.
The tested PGPF isolates produce significant amounts of EPS even at higher NaCl concentration in F36 (3.54 g/ml) followed by F45 (2.1 g/ml) and F62 (1.56 g/ml) isolates; however, EPS production decreases with increasing salt concentration in other tested isolates. EPS constitute a minor fraction (0.1 to 1.5%) of the total soil organic matter. Their unique water-holding and cementing properties play a vital role in the formation and stabilization of soil aggregates and regulation of nutrient and water flow across the plant roots.76 EPS is added to the soil as either sloughed off plant parts and root exudates of intact plants or capsular and slime materials secreted by soil microbes. EPS, or exopolysaccharides, forms a binding association with sodium ions, consequently alleviating the impact of elevated soil salinity. The exopolysaccharides produced by microorganisms play a crucial role in mitigating salt stress by preserving the Na+/K+ balance, thereby aiding plants in surviving adverse soil conditions.77 Osemwegie78 listed different types of EPS produced by common fungal species such as Botryosphaeria sp. (Botryosphaeran) Aureobasidium pullulans (Pullulan), Schizophyllum commune (Schizophyllan), and Aspergillus fumigatus (Galactosaminoglucan). Abbas46 reported that the EPS produced by PGP rhizobacteria have a significant role in plant growth during salinity stress conditions; they achieve this by producing hydrophilic compounds; in addition, they are involved in formation of a coating around the rhizosphere root.
Molecular characterization of selected PGPF
The isolates possessed significant salt tolerance; the PGPF activity under maximum salinity was identified using 18S rRNA sequencing for the identification of fungi and identified as Penicillium citrinum (MF170859), Trichoderma viride F1 (MF170862), Trichoderma viride F2 (MF170858), Acremonium borodinense (MF170860), and Periconia variicolor (MF170861). To the best of our knowledge, this is the first report on the isolation of salt-tolerant PGPF from the rhizosphere of agroforestry tree species. Nazareth and Marbaniang79 isolated a halotolerant Penicillium sp. from mangroves and salterns, recorded their resistance towards heavy metals. All Penicillia showed extreme halo tolerance, and they are able to grow in the absence of salt as well. Plants inoculated with Trichoderma isolates exhibit tolerance against biotic and abiotic stresses, through increased nutritional uptake, enhanced root length, and induced protection against oxidative stress.80 Adedayo and Babalola81 explained the importance of plant growth promoting fungi such as Aspergillus flavus, Actinomucor elegans, Gliocladium virens, Penicillium digitatum, Podospora bulbillosa, Trichoderma sp., and Arbuscular mycorrhizal fungi; they are ecofriendly and enhance the yield of crops through promoting the growth of the roots and shoots and increasing the germination of seeds and the production of chlorophyll for photosynthesis, resulting in high crop yields. Dukare82 reported the plant growth promoting traits of the rhizosphere fungi, Trichoderma species (T. viride, T. harzianum) and Penicillium chrysogenum. Plant cell walls are stimulated by PGPF; this helps avoid nutrient loss during abiotic stress conditions.83
Conclusion
The global population is estimated to be 7.8 billion. This number is expected to rise in to 9.7 billion by 2050. More than half of the lands are affected by salinity typically due to secondary salination, and most of these lands are not suitable for agriculture production. Agroforestry is one of the important alternative income sources for farmers, who own salt-affected lands in the study locations. Agroforestry can improve and restore soil quality and bio-diversity, enhance nutrient cycling, increase carbon sequestration, and reduce climate pressure in degraded lands. With increasing demand for industrial wood, fuel, and timber, plantation forestry, especially those outside the forest area has gained importance. The high salinity and low soil fertility in tropical regions results in poor plant growth and mortality of newly-transplanted tree seedlings. PGP microbes are bio-inoculants, which are promising in forestry for increasing the survival and growth of tree species in saline lands and in other problematic soils.
In this study, we aimed to isolate and identify the native salt-tolerant PGPF from agroforestry plantations in saline soils at different study sites. However, there was only limited data on the presence of free-living rhizosphere soil fungi. We isolated five promising salt-tolerant PGPF and evaluated the possibility of their application as potential bioinoculants that could be cost-effective, more economical than the application of synthetic chemical fertilizers. Once these stress-tolerant bio-inoculants are established in the roots of selected tree seedlings, they can further multiply in the roots and supply the essential nutrients to the host plants in the salt-affected areas; in addition, they can help plants survive better in stressed areas. This study provides valuable insights into the diversity status of salt-tolerant fungal species from agroforestry areas in Karur district; however, it has certain limitations associated with sample collection. The samples were collected only from one particular district and the sample size was small. These limitations highlight the need for further research using a larger sample size to enhance the validity and generalizability of the findings. By addressing these limitations, future studies can contribute to a more comprehensive understanding of the large diversity of fungal isolates and their PGP activity.
Acknowledgments
We thank the Director, Institute of Forest Genetics and Tree Breeding, Coimbatore for providing lab facilities to perform the fungal culture experiments. We also thank the Principal, Government Arts and Science College Kozhinjampara for continuous motivation to complete the work and all the supports towards publishing this work.
Funding Sources
There is no funding sources.
Conflict of Interest
Authors have declared that no competing interests exist.
Authors’ Contribution
Saranya Devi K – analysis and interpretation of results, manuscript preparation.
Mohan V- study conception and design, final review of manuscript.
Data Availability Statement
The manuscript incorporates all datasets produced or examined throughout this research study.
Ethics Approval Statement
Not applicable to this research.
References
- Biswas, Biswas A. Comprehensive approaches in rehabilatingsalt affected soils: A review on Indian perspective. Open Transcation on Geoscience. 2014; 1: 13–24. https://doi.org/10.15764/GEOS.2014.01003
CrossRef - Berg, Zachow C., Muller H., Philipps J., Tilcher R. Next generation bio-products sowing the seed of success for sustainable agriculture. Agronomy. 2013; 3(4): 648–656. https://doi.org/10.3390/agronomy3040648
CrossRef - Qadir M., Oster J.D. Crop and irrigation management strategies for saline-sodic soils and waters aimed at environmentally sustainable agriculture. Sci Total Environ. 2004; 323(1-3):1-19. https://doi.org/10.1016/j.scitotenv.2003.10.012
CrossRef - Banyal, Rajkumar M. K., Rajender K. Y. Agroforestry for rehabilitation and sustenance of saline ecologies. Agroforestry.2017; 413–454.
CrossRef - Meignanamoorthi A., Satheeshkumar S. Impacts of long term irrigation of treated paper mill effluent on groundwater in Karur block. International journal of engineering research and technology. 2018; 6(10): 1-10. https://doi.org/10.17577/IJERTCONV6IS10004
- Arumugam B., Udayasoorian C., Shanmugam T.R., Jayabalakrishnan R. M. Kumar K. Environmental and socio-economic impact of treated paper mill effluent irrigation in Karur district of Tamil Nadu on food safety and food security. First Agricultural Graduate Student Conference 2013 on Food Safety and Food Security At: TNAU, Coimbatore. Madras Agric. J. 2013; 336-342.
- Motos R. A., Ortuno M. F., Vicente A. B., Pedro D.V., Maria J. S. B., Jose A. H. Plant response to salt stress; Adaptive mechanisms. Agronomy. 2017; 7(18): 3–38. https://doi.org/10.3390/agronomy7010018
CrossRef - Wahid A., Perveena M., Gelania S., Basra S.M.A. Pretreatment of seed with H2O2improves salt tolerance of wheat seedlings by alleviation of oxidative damage and expression of stress proteins. Journal of Plant Physiology. 2007;164: 283-294. https://doi.org/10.1016/j.jplph.2006.01.005
CrossRef - Panta, Flowers T. J., Lane P., Doyle R., Haros G., Shabala S. Halophyte agriculture: Success stories. Environmental and Experimental Botany. 2014; 107: 71–83. https://doi.org/10.1016/j.envexpbot.2014.05.006
CrossRef - Munns, Tester M. Mechanism of salinity tolerance. Annual Reviewof Plant Biology. 2008; 59: 651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
CrossRef - Gamalero, Berta G., Massa N., GlickB. R., Lingua G. Interactions between Pseudomonas putida UW4 and Gigaspoa rosea BEG9 and their consequence for the growth of cucumber under salt stress condition. Journal of Applied Microbiology. 2010; 108(1): 236–245. https://doi.org/10.1111/j.1365-2672.2009.04414.x
CrossRef - Ramadoss, Lakkineni V. K., Bose P., Ali S., Annapurna K. Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. SpringerPlus.2013; 2(1): 6. https://doi.org/10.1186/2193-1801-2-6
CrossRef - Naziya, Murali M., Amruthesh K. N. Plant Growth-Promoting Fungi (PGPF) instigate plant growth and induce disease resistance in Capsicum annuum L. upon infection with Colletotrichum capsici (Syd.) Butl. Bisby BioMol. 2020; 10: 41. https://doi.org/10.3390/biom10010041
CrossRef - Egamberdieva D., Wirth S., Bellingrath-Kimura S.D., Mishra J., Arora N.K. Salt-tolerant plant growth promoting rhizobacteria for enhancing crop productivity of saline soils. Front. Microbiol. 2019; 10: 2791. https://doi: 10.3389/fmicb.2019.02791
CrossRef - Dandwate S. C., Analysis of soil samples for its physicochemical parameters from Sangamner city. GSC Biological and Pharmaceutical Sciences. 2020; 12(2): 123-128. https://doi.org/10.30574/gscbps.2020.12.2.0243
CrossRef - Jayaraman P., Shalini S., Saraswathi K., Vadamalai K., Logambal R. Comparative Studies on Fungal Biodiversity of agricultural field soil from Thiruvannamalai District, Tamil Nadu, India. Int. J. Curr.Microbiol. App. Sci. 2018; 7(7): 4259-4273. https://doi.org/10.20546/ijcmas.2018.707.497
CrossRef - Ji C., Wang X., Song X., Zhou Q., Li C., Chen Z., Gao Q., Li H., Li J. Zhang P., Cao H. Effect of Bacillus velezensisJC-K3 on endophytic bacterial and fungal diversity in wheat under salt stress. Front. Microbiol. 2021; 12. https://doi: 10.3389/fmicb.2021.802054
CrossRef - TamieAl S. S. A. Effect of salinity on the fungal occurrence in Al-shega area at Al-Qassim, Saudi Arabia. Research Journalof Microbiology. 2014; 9(6): 287–295. https://doi.org/10.3923/jm.2014.287.295
CrossRef - Sharma, Gerdemann K. G., Agrawal A., Bhatnagar M., Sharma R. Effect of phosphate solubilizing bacteria on the germination of Cicer arietinumseeds and seedling growth. Herb, J. Med. and Toxicol. 2007; 1: 61–63.
- Devendra J., Jyoti S., Gunnjeet K., Ali Asger B., Surya C.,Vimal S., Archna S., Santosh Ranjan M., Elina M. Phenetic and molecular diversity of nitrogen fixating plant growth promoting Azotobacter isolated from semiarid regions of India. BioMed Research International. 2021; 9. https://doi.org/10.1155/2021/6686283
CrossRef - Hofmann M., Heine T., Malik L., Hofmann S., Joffroy K., Senges C.H.R., Bandow J.E., Tischler D. Screening for microbial metal-chelating siderophores for the removal of metal ions from solutions. Microorganisms. 2021; 9(1):111. https://doi.org/10.3390/microorganisms9010111
CrossRef - Joseph, Patra R. R., Lawrence R. Characterization of plant growth promoting rhizobacteria associated with chickpea(Cicer arietinum L.). International Journal of Plant Production. 2007; 2: 141–152. https://doi.org/10.22069/IJPP.2012.532
- Penrose M., Glick B. R. Methods for isolating and characterizing ACCdeaminase containing plant growth-promoting rhizobacteria. Physiologia Plantarum. 2003; 118(1): 10–15. https://doi.org/10.1034/j.1399-3054.2003.00086.x
CrossRef - Turbat A., Rakk D., Vigneshwari A., Kocsube S., Thu H., Szepesi A., Bakacsy L.D., Skrbic B., Jigjiddorj E.A., Vagvolgyi C. Characterization of the plant growth-promoting activities of endophytic fungi isolated from Sophora flavescens. Microorganisms. 8(5):683. https://doi.org/10.3390/microorganisms8050683
CrossRef - Bano N., Musarrat J.Characterization of a new Pseudomonas aeruginosa strain NJ-15 as a potential bio-control agent. Current Microbiology. 2003; 46(5): 324–328. https://doi.org/10.1007/s00284-002-3857-8
CrossRef - Ruhmann B., Schmid J., Sieber V. Methods to identify the unexplored diversity of microbial exopolysaccharides. Front. Microbiol. 2015; 6: 565. doi: 10.3389/fmicb.2015.00565
CrossRef - Wang Z., Wang Y., Dong Q., Fan Q., Dao V.M., Yu H. Morphological and phylogenetic characterization reveals five new species of Samsoniella(Cordycipitaceae, Hypocreales). J Fungi (Basel). 2022; 19: 8(7):747. doi: 10.3390/jof8070747
CrossRef - Hunt, Boddy L., Randerson P. F., Rogers H. J.A evaluation of 18S rDNA approaches for the study of fungal diversity in grassland soils. Microbial Ecology. 2004; 47(4): 385–395. https://doi.org/10.1007/s00248-003-2018-3
CrossRef - Kaur M., Li J., Zhang P., Yang H.F., Wang L., Xu M. Agricultural soil physico-chemical parameters and microbial abundance and diversity under long-run farming practices: A greenhouse study. Front. Ecol. Evol. 2022; 10. doi: 10.3389/fevo.2022.1026771
CrossRef - Rousk, Brookes P. C., Baath E.Contrasting soil pH effects on fungal and bacterial growth suggests functional redundancy in carbon mineralisation. Applied and Environment Microbiology. 2009; 75: 1589–1596. https://doi.org/10.1128/AEM.02775-08
CrossRef - Iwai B., Oo A. N., Topark-ngarm B. Soil property and microbial activity in natural salt affected soils in an alternating wet–dry tropical climate. Geoderma. 2012; 189–190: 144–152. https://doi.org/10.1016/j.geoderma.2012.05.001
CrossRef - Sharma S., Totat K. L., Shyampur R. L. Characterization and classification of salt affected soils of southern Rajasthan. Journal of the Indian Society of Soil Science. 2004; 52(3): 209–214.
- Kawser U., Nath B., Hoque A. Observing the influences of climatic and environmental variability over soil salinity changes in the Noakhali coastal regions of Bangladesh using geospatial and statistical techniques. Chall.2022; 6: 100429. https://doi.org/10.1016/j.envc.2021.100429
CrossRef - Klein, Stewart J.D., Porter S. S., Weedon J. T., Kiers E. T. Evolution of manipulative microbial behaviours in the rhizosphere. Evolutionary Applications. 2022; 15(10): 1521–1536. https://doi.org/10.1111/eva.13333
CrossRef - Msimbira L. A., Smith D. L. The roles of plant growth promoting microbes in enhancing plant tolerance to acidity and alkalinity stresses. Front. Sustain. Food Syst. 2020; 4:106. https://doi: 10.3389/fsufs.2020.00106
CrossRef - Ponmurugan P., Gopi C. G. Distribution pattern and screening of phosphate solubilizing bacteria isolated from different food and forage crops. Journal of Agronomy. 2006; 5(4): 600–604. https://doi.org/10.3923/ja.2006.600.604
CrossRef - Grishkan I., Nevo E., Wasser S. P. Soil micromycete diversity in the hypersaline dead sea coastal area, Israel. Mycological Progress. 2003; 2(1): 19–28. https://doi.org/10.1007/s11557-006-0040-9.
CrossRef - Ibekwe M., Poss J. A., Grattan S. R., Grieve C. M., Suarez D. Bacterial diversity in cucumber (Cucumis sativus) rhizosphere in response to salinity, soil pH, and boron. Soil Biologyand Biochemistry. 2010; 42(4): 567 575. https://doi.org/10.1016/j.soilbio.2009.11.033
CrossRef - Zhang K., Shi Y., Cui X., Yue P., Li K., Liu X., Tripathi, B. M., Chu H. Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems. 2019; 12; 4(1): e00225-18. https://doi: 10.1128/mSystems.00225-18.
CrossRef - Dave S., Desai B. Microbial diversity at marine salterns near Bhavnagar. Current Science. 2006; 90 (4): 497-500.
- Oren, A. Molecular ecology of extremely halophilic archaea and bacteria. FEMS Microbiol. Ecol.2002, 39, 1–7. 1111/j.1574-6941.2002.tb00900.x
CrossRef - Mayak, Tirosh T., Glick B. R. Plant growth promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiology and Biochemistry. 2004; 42(6): 565–572. https://doi.org/10.1016/j.plaphy.2004.05.009
CrossRef - Choudhary KK. Occurrence of nitrogenfixing cyanobacteria during different stages of paddy cultivation. Bangladesh Journal of Plant Taxonomy. 2011; 18(1): 73–76. http://doi.org/3329/bjpt.v18i1.7842
CrossRef - Arit, Leon D.L., Laura D. B., Luc D. Soil salinity controls relative abundance of specific bacterial groups involved in decomposition of maize plant residues. Frontiers in Ecology and Evolution. 2018; 14. https://doi.org/10.3389/fevo.2018.00051
CrossRef - Mohan, Saranya DeviK. Selection of potential isolates of Plant Growth Promoting Rhizobacteria (PGPR) in conferring salt tolerance under in vitro. Journal of Academia and Industrial Research. 2015; 4(3): 91–99.
- Abbas, Rasul S., Aslam K., Baber M., Shahid M., Fathia M., Tahir N. Halotolerant PGPR: A hope for cultivation of saline soils. Journal of King Saud University – Science. 2019; 31(4): 1195–1201. https://doi.org/10.1016/j.jksus.2019.02.019.
CrossRef - Srividya, Soumya S., Pooja K. Influence of environmental factors and salinity on P solubilization by a newly isolated Aspergillus nigerF from agricultural soil. Afr. J of Biotechnol. 2009; 8(9): 4.
- Malgioglio, Rizzo G. F., Nigro S., Lefebvre Du Prey V., Herforth-Rahme J., Catara V., Branca F. Plant-microbe interaction in sustainable agriculture: The factors that may influence the efficacy of PGPM application. Sustainability. 2022; 14(4): 2253. https://doi.org/10.3390/su14042253
CrossRef - Bhattacharya, Bachani P., Jain D., Patidar S. K. Exraction of potassium from K-feldspar through potassium solubilization in the halophilic Acinetobacter soil isolated from experimental salt farm. International Journal of Mineral Processing. 2016;152: 53–57. https://doi.org/10.1016/ j.minpro.2016.05.003
CrossRef - Parmar P., SindhuS. Potassium solubilization by rhizosphere bacteria: Influence of nutritional and environmental conditions. Journal of Microbiology Research. 2013; 3 (1): 25–31. https://doi.org/10.5923/j.microbiology.20130301.04
- Etesami, Emami S., Ali H. A.Potassium solubilising bacteria. Journal of Soil Science and Plant Nutrition. 2017; 17 ( 4): 897-911.
CrossRef - Tavallali, Rahemi M., Eshgi S., Ramezanian. Zinc alleviates salt stress and increases antioxidants enzyme activity in the leaves of Pistachioseedling. Truk. J. gr forest. 2010; 34(4): 349–359. https://doi.org/10.3906/tar-0905-10
CrossRef - Saravanan S., Kumar, M. R., Sa T. Microbial zinc solubilization and their role of plants. Bacteria in Agrobiology. Plant nutrient management. 2011; 1: 47–63. https://doi.org/10.1007/978-3-642-21061-7_3.
CrossRef - Paul, Rakshit A. Evaluating lignification, antioxidativedefense, and physiochemical changes in soybean through bio-priming under graded soil fertilization. Journal of Soil Science and Plant Nutrition. 2022; 22(2): 2295–2306. https://doi.org/10.1007/s42729-022-00809-9
CrossRef - Anand A,, Chinchilla D,, Tan C,, Mene-Saffrane L., L’Haridon F., Weisskopf L. Contribution of hydrogen cyanide to the antagonistic activity of Pseudomonasstrains against Phytophthora infestans . Microorganisms. 2020; 8(8):1144. https://doi.org/10.3390/microorganisms8081144
CrossRef - Rijavec T., Lapanje A. Hydrogen cyanide in the rhizosphere: Not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 2016; 7:1785. doi: 10.3389/fmicb.2016.01785
CrossRef - Fairbrother, Shapter J., Brugger J., Southam G., Pring A., Reith F. Effect of the cyanide-producing bacterium Chromobacterium violaceumon ultraflat Au surfaces. Chemical Geology. 2009; 265(3–4): 313–320. https://doi.org/10.1016/j.chemgeo.2009.04.010
CrossRef - Lapanje, Wimmersberger C., Furrer G., Brunner I., Frey B. Pattern of elemental release during the granite dissolution can be changed by aerobic heterotrophic bacterial strains isolated from Damma glacier (Central Alps) deglaciated granite sand. MicrobialEcology. 2012; 63(4): 865–882. https://doi.org/10.1007/s00248-011-9976-7
CrossRef - Wongfun, Plotze M., Furrer G., Brandl H. Weathering of granite from the Damma glacier area: Thecontribution of cyanogenic bacteria. Geomicrobiology Journal. 2014; 31(2): 93–100. https://doi.org/10.1080/01490451.2013.802396
CrossRef - Murali M., Naziya B., Ansari M.A., Alomary M.N., Alyahya S., Almatroudi A., Thriveni M.C., Gowtham H.G., Singh S.B., Aiyaz M. Bioprospecting of rhizosphere-resident fungi: Their role and importance in sustainable agriculture. Journal of Fungi. 2021; 7(4):314. https://doi.org/10.3390/jof7040314
CrossRef - Contreras-Cornejo A., Macias-Rodríguez L., del-Val E., Larsen J. Ecological functions of Trichoderma spp. and their secondary metabolites in the rhizosphere: Interactions with plants. FEMS Microbiology Ecology. 2016; 92(4): 36. https://doi.org/10.1093/femsec/fiw036
CrossRef - Prasad R., Sagar B. V., Devi G. U., Triveni S., Rao S. R. K., Chari K. D. Isolation and screeningof bacterial and fungal isolates for plant growth promoting properties from tomato (Lycopersicon esculentum Mill.). International Journal of Current Microbiology and Applied Sciences. 2017; 6(8): 753–761. https://doi.org/10.20546/ijcmas.2017.608.096
CrossRef - Salas-Marina A., Silva-Flores M. A., Cervantes-Badillo M. G., Rosales-Saavedra M. T., Islas-Osuna M. A., Casas-Flores S. The plant growth-promoting fungus Aspergillus ustus promotes growth and induces resistance against different lifestyle pathogens in Arabidopsis thaliana. Journal of Microbiology and Biotechnology. 2011; 21(7): 686–696. https://doi.org/10.4014/jmb.1101.01012
CrossRef - Crowley D.E. Iron nutrition in plants and rhizospheric microorganisms. Springer; Dordrecht, The Netherlands. Microbial siderophores in the plant rhizosphere; 2006:169–198.1007/1-4020-4743-6_8
CrossRef - Ghosh K., Banerjee S., SenGupta, C. Bioassay, characterization and estimation of siderophores from some important antagonistic Fungi. Journal of Biopesticides. 2017; 10(2): 105–112. https://doi.org/10.57182/jbiopestic.10.2.105-112
CrossRef - Mahmoud., Esam., Abd-Alla., Howaida. Siderophores production by some microorganisms and their effect on Bradyrhizobium-Mung bean symbiosis. Int. J. Agr. Biol. 2001; 3.
- Zhang S., Gan Y.. Xu B. Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma longibrachiatum T6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biol. 2019; 19: 22. https://doi.org/10.1186/s12870-018-1618-5
CrossRef - Viterbo A., Horwitz A. Mycoparasitism. InK. Ebbole (Ed.), Cellular and molecular biology of filamentous fungi. American Society for Microbiology, 2010; 676–693.
CrossRef - Shaharoona, Arshad M., Zahir Z. A. Effect of plant growth promoting rhizobacteria containing ACC‐deaminase on maize (Zea maysL.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Letters in Applied Microbiology. 2006; 42(2): 155–159. https://doi.org/10.1111/j.1472-765X.2005.01827.x
CrossRef - Safronova I., Stepanok V. V., Engqvist G. L., Alekseyev Y. V., Belimov A. A.Root associated bacteria containing 1-aminocyclopropane-1-carboxylate deaminase improve growth and nutrient uptake by pea genotypes cultivated in cadmium supplemented soil. Biology and Fertility of Soils. 2006; 42(3): 267–272. https://doi.org/10.1007/s00374-005-0024-y
CrossRef - Zhang S., Gan Y., Xu B. Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma longibrachiatumT6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biology. 2019;19:22–22. 10.1186/s12870-018-1618-5.
CrossRef - Egamberdieva D., Wirth S.J., Alqarawi A.A., Abd Allah E.F., Hashem A. Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Front Microbiol. 2017; 31(8):2104. doi: 10.3389/fmicb.2017.02104.
CrossRef - Van Loon C. Plant responses to plant growth-promoting rhizobacteria. European Journal of Plant Pathology. 2007; 119(3): 243–254. https://doi.org/10.1007/s10658-007-9165-1
CrossRef - Deng Z., Cao, L. Fungal endophytes and their interactions with plants in phytoremediation: a review. Chemosphere. 2017; 168: 1100–1106. doi: 10.1016/j.chemosphere.2016.10.097
CrossRef - Muhammad I., Ali N., Jan G., Jan F. G., Rahman I. U., Iqbal, A. IAA producing fungal endophyte Penicillium roqueforti, enhances stress tolerance and nutrients uptake in wheat plants grown on heavy metal contaminated soils. PLoS One. 2018;13:e0208150. doi: 10.1371/journal.pone.0208150
CrossRef - Mahapatra S., Banerjee D. Fungal exopolysaccharide: production, composition and applications. Microbiol Insights. 2013; 29: 6:1-16. doi: 10.4137/MBI.S10957
CrossRef - Singh R.P., Jha P.N. 2016. A halotolerant bacterium Bacilluslicheniformis HSW-16 augments induced systemic tolerance to salt stress in wheat plant (Triticum aestivum). Plant Sci. 7: 1890. doi: 10.3389/fpls.2016.01890
CrossRef - Osemwegie O.O., Adetunji C.O., Ayeni E.A., Adejobi O.I., Arise R.O., Nwonuma C.O., Oghenekaro A.O. Exopolysaccharides from bacteria and fungi: current status and perspectives in Africa. Heliyon. 2020; 15: 6(6). doi: 10.1016/j.heliyon.2020.e04205.
CrossRef - Nazareth S., Marbaniang T. Effect of heavy metals on cultural and morphological growth characteristics of halotolerant Penicilliummorphotypes. Journal of Basic Microbiology. 2008; 48(5): 363–369. https://doi.org/10.1002/jobm.200800006
CrossRef - Akash H. Plant response to Trichodermaspp and their tolerances to biotic stresses. A review. of pharmaco and phytochemistry. 2018; 7(1): 758–766.
- AdedayoA., Babalola O. O. Fungi that promote plant growth in the rhizosphere boost crop growth. Journal of Fungi. 2023; 9(2): 239. https://doi.org/10.3390/jof9020239
CrossRef - Dukare S., Singh R. K., Jangra R. K., Bhushan B. Non fungicides- Based promising technologies for managing postproduction Penicillium induced spoilage in horticultural commodities: A comprehensive review. Food Reviews International. 2022; 38(3): 227–267. https://doi.org/10.1080/87559129.2020.1727497
CrossRef - Koza A., Adedayo A. A., Babalola O. O., KappoA. P. Microorganisms in plant growth and development: Roles in abiotic stress tolerance and secondary metabolites secretion. Microorganisms. 2022; 10(8): 1528. https://doi.org/10.3390/microorganisms10081528
CrossRef

