Introduction
The population of world is increasing day by day. India has reached 1.21 billion, whereas population of Rajasthan is 6.85 crore. In Rajasthan, the average size of operational land holdings is also decreasing and it is 2.73 hectare. It will be a challenging task to elevate the agricultural production whereas rate of population and urbanization is increasing1. To increase the production and economically self-stabilization, farmers use chemical fertilizers in the fields. Human health as well as ecological imbalance is the major concerns in use of these chemical fertilizers. Nature gave us the most efficient and self-governing process called Symbiotic nitrogen fixation to overcome dependency on chemical nitrogen fertilizers2. Legumes have the ability to establish this mutual relationship with soil rhizobia that helps legumes to use environmental nitrogen3-4. Biological nitrogen fixation contributes around 60% of total nitrogen fixation taking place on earth5. Rhizobia represent a diverse group of bacteria that are characterized as gram-negative, rod-shaped, and non-spore-forming. Rhizobia are classified into 18 genera and 238 species of nodulating bacteria belonging to three class alphaproteobacteria, beta proteobacteria and gamma proteobacteria6. The Family leguminoseae is 3rd largest family of angiosperms comprising 751 genera and 19,500 species7. The connection establishment between rhizobia and legumes depends on several environmental abiotic factors like salinity in rhizosphere, soil temperature and draught conditions, which can act as a barrier in nodule formation8-9. Nodules are the favourable spots for bacteria as they provide a protected environment for their multiplication and survival10.
In the Great Thar Desert of India, species of Ensifer are predominant in forming nodules on legume plants, however other nodulating rhizobia were also reported11-15. The Aravalli Range, an ancient mountain range, serves as a natural barrier that prevent the expansion of the Thar Desert. The central part of the Aravalli range extends across several districts in Rajasthan, including Ajmer, Jaipur, and the southwestern portion of Tonk. It comprises around 21000 square km. area. Recently, some legume species in the Central Aravalli region have been studied for their associated rhizobia16-17. Yet, many species of nodulating legumes in this area have not been characterized in terms of their microsymbionts. The current study is aimed at identifying and characterizing the rhizobia associated with the Zornia gibbosa Span. Native legume Zornia gibbosa is a branched, prostate, diffused of sub-erect annual herb with raceme inflorescence and yellow flowers. The flowering and fruiting occur in the month of August to October during monsoon season. It is commonly found in wastelands and grass lands with wet, shaded habitat18. Characterizing the rhizobia associated with indigenous legumes enhances our knowledge of the symbiotic interactions between legumes and rhizobia. This research provides important insights into their ecological functions and the potential advantages they offer for sustainable agricultural practices in the region.
Material and methods
Survey and collection
Root nodules of Zornia gibbosa were collected during monsoon season from various sites at Jaipur (Jhalana, Amer, Jamwaramgarh and Achrol) and Ajmer (MDSU Campus, Pushkar, Picholiya) districts in Central Aravali range. Viable seeds and rhizospheric soil were also collected from these sites for rhizobia trapping experiment.
Rhizobia trapping experiment & Nodule harvestation
Healthy Seeds of Zornia gibbosa were surface sterilized followed by scarification and dipped into water for about an hour. Swollen seeds were sowed in pots containing rhizospheric soil during monsoon season. After 8 weeks the healthy plants were harvested for their nodules by using standard methods19.
Isolation and purification
With the help of scalpel blade nodules were excised from the roots and surface sterilized with 90% ethanol for half minute, then 0.1% mercuric chloride for 3 min. and followed by six rinses with sterile distilled water as described in standard protocol19. Congo red-Yeast extract mannitol agar (CR-YEMA) plates were prepared for isolation and purification of root nodule bacteria20. Surface sterilized nodules were crushed on sterile plates and loop filled with nodule suspension streaked on CR-YEMA plates and plates were incubated at 28°C for 48-72 hours. Plates having more than one type of colonies were further processed to obtain separate colonies and re-streaked to get pure culture. Pure isolates were given a name and stored at 4°C.
Phenotypic and Biochemical Characterization:
All purified isolates were tested for following phenotypic and biochemical parameters by using standard methods and protocols.
pH tolerance
YEMA plates with the range of pH 5 to 10 were inoculated with pure bacterial culture and incubated at 28°C for 48-72 hours and recoded for tolerance to acidic or alkaline pH. The pH was adjusted with the help of 1N NaOH and 1N HCI21. Growth on various pH will be considered as positive result.
NaCl tolerance
With the salt concentration of 0.5 to 6% (w/v) range, isolates were tested for NaCl tolerance. YEMA plates with above salt concentration were inoculated with bacterial culture and incubated at 28°C for 48-72 hours. The bacterial growths on plates were noted as positive results that indicate the ability of isolates to withstand the respective concentration of salt, thus providing a measure of its salt tolerance capability.
BTB Reaction
The alkali or acid production by the bacterial isolates was determined by bromo thymol blue (BTB) reaction. For this BTB-YEM broth19 is prepared and inoculated with fresh activated rhizobial cultures. After inoculation, incubated in shaking incubator at 30°C and 100 rpm for 2-3 days. The colour of control BTB-YEM broth was green at neutral pH. Acid producing bacteria turned green colour in yellow colour whereas alkali producing turned in blue colour.
Oxidase activity
To determine the oxidase activity in rhizobial strains, white oxidase test discs were placed onto fresh cultures of these strains. A change in color of the discs from white to blue signifies the presence of oxidase activity in the rhizobia, indicating a positive result. Conversely, if the disc retains its original white color, it is indicative of a negative result, showing the absence of oxidase activity.
Citrate utilization
Simmons citrate agar is used to detect the ability of bacteria to utilize citrate as source of carbon21. For this Simmons citrate agar slants were formed and then inoculated with fresh activated bacterial strains and then incubated at 28°C for 2-3 days. Change in the colour of slants from green to blue indicates that bacteria use citrate as an energy or carbon source, while no change in colour shows negative result.
Catalase test
To confirm the catalase enzyme activity of rhozobial strains, test tubes containing fresh YEM broth medium were inoculated with fresh rhizobial strains and incubated at 28°C for 3-4 days. After incubation 3-4 drops of 3% hydrogen peroxide (H2O2) were added in each culture tubes. Release of oxygen gas due to breakdown of H2O2, leads to bubbling or foaming in test tubes and considered as positive result21.
Cellulase activity
The ability of rhizobial isolates to produce extracellular cellulase enzymes enables them to catalyze the breakdown of cellulose into sugars. To perform this test Carboxy methyl cellulose (CMC) Congo red plates were inoculated with activated rhizobial cultures and incubated at 28°C for 3-5 days. Cellulase producing bacteria used this CMC as substrate and showed halo zone around their colony and no halo zone represented negative results.
Gelatin hydrolysis
This test detects the activity of gelatinase enzyme that rapidly hydrolyses the gelatin and to use gelatin as a source of nutrient by rhizobia. For this, nutrient gelatin media (peptone water with 12% gelatin) containing tubes were inoculated by fresh rhizobial culture and incubated at 28°C and 120 rpm for 3-5 days. After incubation tubes were kept in refrigerator at 4°C for 30 min. to solidify the media. Inoculated tubes that remained in liquid state considered as positive result (activity of Gelatinase in hydrolysis) while solidification considered as negative results. Re-incubate all the solidified tubes for additional 5 days and then again kept at 4°C for 30 min. liquefied tubes showed slow gelatin hydrolysis21.
Amylase production
The amylase enzyme catalyzes the hydrolysis of starch into simpler sugars. To assess the capability of rhizobia to utilize starch as a carbon source, a specific test was conducted. Fresh inoculums of rhizobia were spread on nutrient agar plates that were enriched with 0.2% soluble starch. These plates were then incubated at a temperature of 28°C for a period ranging from 24 to 72 hours. Amylase converts starch into sugar by hydrolysis. Amylase producing ability of rhizobia were accessed by inoculating fresh rhizobial cultures onto nutrient agar medium plates supplemented with 0.2% soluble starch. Inoculated plates were incubated at 28°C for 24-72 hours and subsequently these plates were flooded with iodine solution. A clear halo zone around colonies (due to starch hydrolysis) considered as positive result21.
Protease activity
Proteolytic enzymes (proteases) convert complex protein casein (milk protein), into its simple forms like polypeptides, dipeptides and up to individual amino acids. To perform this test skim milk agar plates were inoculated with fresh rhizobial strains and incubated at 28°C for 24-72 hours. A clear halo proteolysis zone around colonies indicates secretion of protease by particular isolate and considered as positive result.
Indole production
This determine whether rhizobial strains are able to convert tryptophan into the indole or not. Test tubes filled with 1% tryptone broth were inoculated with fresh inoculum and incubated at 28°C for 48-72 hours. Kovac’s reagent was further added to culture tubes and pink layer on the top of the media considered as positive result for indole production.
Results and Discussion
Root nodules from the leguminous plant Zornia gibbosa were gathered across eight locations within the Central Aravalli range, including Jhalana, Amer, Jamwaramgarh, Achrol in Jaipur, as well as the MDSU Campus, Pushkar, and Picholiya in Ajmer (Figure 1). Additionally, nodules were also collected from rhizobia trapping experiments, enriching the diversity for study. The field view of plant along with their flowers and pods, the setup of the rhizobia trapping experiment, and the plant’s complete root system with nodules were shown in figure 2A-E. The nodules in Zornia gibbosa showed a pattern of determinate growth, took on a globular shape, and were predominantly found on the plant’s primary roots. A similar type of nodules has been reported in Alysicarpus and Rhynchosia in Thar Desert11. These nodules were further processed to get root nodule isolate on YEMA culture plates. A total of 44 isolates of RNB (root nodule bacteria) were purified and examined for their colony characteristics. Most of the RNB isolates were showed rhizobia like colony characteristics like white, translucent/opaque, mucilaginous, raised, entire with smooth margins (Table 1, Fig 2F). Similar observation also reported by other researchers12-17, 19.
 |
Figure 1: Root nodules and germplasm collection sites in Central Aravalli range |
 |
Figure 2: Plant system Zornia gibbosa: field view of plant (A), flowers and pods (B), rhizobia trapping experiment (C), root system with nodules (D-E) and pure culture of rhizobia (F) |
Table 1. Colony characteristics of RNB strains isolated from Zornia gibbosa
| S. No | Colony characteristics | Isolate |
| 1. | White, opaque, non-mucilaginous, raised, non-gummy, smooth margins | Z1, Z2, Z4, Z11, Z13 |
| 2. | Watery, transparent, mucilaginous, raised, gummy, smooth margins | Z5, Z7, Z9, Z23 |
| 3. | White, translucent, non-mucilaginous, raised, non-gummy, smooth margins, became dried in some days | Z12, Z22, Z27, Z28, Z30 |
| 4. | Watery, translucent, mucilaginous, raised, gummy, smooth margins | Z3, Z8, Z19 |
| 5. | Watery, translucent, non-mucilaginous, raised, non-gummy, smooth margins | Z6, Z33 |
| 6. | White, translucent, mucilaginous, raised, non-gummy, smooth margins | Z10 |
| 7. | Yellowish white, translucent to opaque, mucilaginous, raised, non-gummy, smooth margins, dried in some days | Z15 |
| 8. | Reddish white, opaque, mucilaginous, raised, non-gummy, smooth margins | Z16, Z17, Z18, Z24, Z31, Z34, Z35, Z44, Z37 |
| 9. | Reddish, opaque, non-mucilaginous, raised, gummy, smooth margins | Z21, Z14 |
| 10. | Reddish watery, translucent, mucilaginous, raised, non-gummy, smooth margins | Z25, Z26, Z38 |
| 11. | Watery, translucent, mucilaginous, raised, non-gummy, smooth margins | Z29 |
| 12. | Reddish white, opaque, mucilaginous, raised, gummy, Smooth margins | Z32 |
| 13. | Reddish watery, translucent to transparent, raised, gummy, smooth margins | Z20, Z36, Z39, Z40, Z41, Z42, Z43 |
All 44 RNB isolates were showed significant diversity in their phenotypic and biochemical characteristics, which are detailed in Table 2 and illustrated in Figure 3. On the basis of BTB reaction tested bacteria were classified as alkali producer or acid producer. Among 44 isolates tested isolates, 8 isolates were acid producing bacteria (appearance of yellow colour) whereas 8 were alkali producers (appearance of blue colour). These results show resemblance with fast and slow growing strains isolated from root nodules of soybean in Brazil22. Similar results were also observed for rhizobial strains isolated from chickpea grown in Ethiopian region23. All isolates were examined for NaCl tolerance with range of 0.5% to 5% NaCl concentration. Most of the strains were restricted upto 3% concentration however Z2, Z13, Z14, Z15, Z21 and Z22 were tolerant upto 5% of NaCl concentration (Table 2, Fig 3). Similar outcomes were also observed by many other reasearchers24-25. Additionally, rhizobial strains isolated from cowpea in Western Kenya were also highly tolerant to NaCl concentrations26. Numerous studies have indicated that rhizobia with salt tolerance capabilities play a significant role in enhancing the ability of associated legume crops to withstand saline conditions27-28. This synergy between salt-tolerant rhizobia and legume crops holds potential for sustainable agricultural practices in saline-affected areas.
All 44 isolates were also tested for their tolerance for pH range of 5 to 10 and distributed in seven groups. Major group comprising twenty-nine strains were able to grow pH range of 5 to 10, while some strains like Z1, Z11, Z29, Z31, and Z33, exhibited heightened sensitivity to changes in pH and were able to grow pH range 7-8 (Table 2, Fig 3). In compare to fast growing bacteria, slow growing bacteria show much tolerance to low pH29. Similarly in present study the slow grower strains (Z3, Z26 and Z32) were showed good growth at pH 5 in compare to other strains. The overall findings indicate that the rhizobia present in the Central Aravalli region are more adapted to alkaline conditions. This adaptation suggests that these microbial populations have evolved mechanisms to thrive in environments with higher pH levels, which is characteristic of the soil conditions in this geographical area. Such a propensity for alkaline tolerance underscores the potential of these rhizobia to support leguminous plant growth in similar soil conditions, highlighting their importance for agricultural practices in the Central Aravalli region and other areas with alkaline soils. Similar results were shared by other researchers in Rajasthan as well as from different country12-14, 30-31.
 |
Table 2: Biochemical characteristics of RNB strains isolated from Zornia gibbosa |
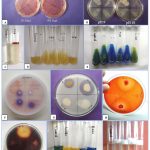 |
Figure 3: Phenotypic and biochemical activities of isolates: (A) NaCl tolerance, (B) pH tolerance, (C) Indole production, (D) Gelatin hydrolysis, (E) Citrate utilization, |
In gelatinase activity all isolates showed negative result except Z2, Z14, Z15, Z32, Z34, Z37, Z42, and Z44 (Table 2). Similar to our findings the rhizobia isolated from Phaseolus vulgaris (Odisha)32, Cajanus cajan (Telangana)33, Cicer arietinum (Afghanistan)34 and Groundnut (Myanmar)35 were also showed negative results for gelatinse activity. In our study 15 isolates were tested positive for catalase activity and 32 isolates were for oxidase activity (Table 2, Fig 3). The catalase activity of rhizobia helps the leguminous plants to mitigate oxidative stress, leading to enhanced growth, increased resilience to environmental stresses, and overall better plant health. Two isolates Z13 and Z24 were found with maximum catalase activity. In present study among 44 RNB isolates only few were showed positive results for citrate utilization (Z4, Z18, Z22, Z32, Z44) and amylase production (Z4, Z6, Z21, Z25, Z42) (Table 2, Fig 3). Similar results were also observed by many other researchers 34, 36-38. In Ethiopian region 92% isolates form Cicer arietinum L. and 100% isolates isolated from rhizosphere of chickpea and pea nodules exhibited amylase production39-40. Bacterial strains exhibiting indole production are recognized as good plant growth-promoting isolates. In our study one isolate Z22 tested positive for indole production (Table 2). Similar observation has been also observed for the rhizobial strains isolated from cowpea, elephant, lablab and mung bean41-42. Out of 44 RNB isolates, 15 isolates were found positive for protease activity (Table 2) and similar results were also recorded from Central Java where 9 isolates showed protease activity43. In present study, we observed positive cellulase activity in 12 isolates. Similarly, 50% RNB isolates of Alysicarpus vaginalis from Thar Desert showed positive results for cellulase activity44. In Ethiopian region 38% and 48% RNB strains isolated from soybean and groundnut respectively, were tested positive for cellulase activity45-46. Over all root nodule bacterial strains associated with Zornia giobbsa were highly diverse in their phenotypic and biochemical characteristics. Certain superior strains identified in this study, such as Z22, Z42 and Z44, which displayed diverse enzymatic activities, hold potential for application in the synthesis of biofertilizers and implementation in revegetation programs.
Conclusion
In the present study, forty-four isolates were isolated from the root nodules of Zornia gibbosa collected from various sites within the Central Aravalli range. All the isolates were tested for their phenotypic and biochemical characteristics. Most of the strains produced white, raised, translucent, raised colonies with smooth margins. The isolates demonstrated tolerance to both salt and pH stress and exhibited positive results for various enzymatic activities, including catalase activity, oxidase activity, protease activity, cellulose activity, BTB reaction, and amylase production. The comprehensive analysis of root nodule bacterial strains associated with Zornia gibbosa revealed a remarkable degree of diversity in both their phenotypic and biochemical features. This significant variation underscores the heterogeneous nature of the root nodule bacterial community associated with Zornia gibbosa, reflecting adaptability and versatility among these bacterial strains in response to the plant’s root nodule environment. Further molecular analysis of selected isolates is needed for their identification and beneficial use in agriculture system.
Acknowledgement
The authors express their sincere gratitude to Rashtriya Uchchattar Shiksha Abhiyan 2.0 (RUSA 2.0) for fortifying and offering essential research facilities in the Department of Botany, University of Rajasthan, Jaipur. Additionally, Ghanshyam Sharma acknowledges the Council of Scientific & Industrial Research (CSIR), New Delhi, for their financial support through fellowships.
Funding source
This work has been supported by the Science and Engineering Research Board (SERB), Government of India, under Grant No. SUR/2022/001518. The Department of Botany gratefully acknowledges the facilities and grants provided by SERB, which have significantly contributed to the success of this work.
Conflicts of interests
The authors report no conflicts of interest concerning the research, authorship, and publication of this article.
Authors contribution
Ghanshyam Sharma: He conducted survey in the Central Aravalli Region, executed trapping experiments, collected nodules, and performed the isolation and characterization of all bacterial isolates. He drafted the manuscript, ensuring effective communication of the study’s objectives, methodologies, results, and conclusions.
Amit Yadav: He conducted data analysis, offered interpretation, and provided an effective discussion.
Sonam Choudhary: She conducted various laboratory experiments as part of this study.
Indu Singh Sankhla: He provided guidance for the study and made necessary corrections in the manuscript.
Data availability
The manuscript incorporates all datasets produced in the study.
Ethics approval and Informed consent statement
Not applicable
References
- Hossain A., Krupnik T. J., Timsina J., Mahboob M. G., Chaki A. K., Farooq M., Bhatt R., Fahad S., Hasanuzzaman M. Agricultural land degradation: processes and problems undermining future food security. In: Fahad S., Hasanuzzaman M., Alam M., Ullah H., Saeed M., Khan I. A., Adnan M. Environment, climate, plant and vegetation growth. Cham: Springer International Publishing; 2020:17-61.
CrossRef - Gauri, Singh A. K., Bhatt R. P., Pant S., Bedi M. K., Naglot A. Characterization of Rhizobium isolated from root nodules of Trifolium alexandrinum. J Agri Technol. 2011;7(6):1705-1723.
- Oldroyd G. E., Downie J. A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59(1):519-546.
CrossRef - Oldroyd G. E., Murray J. D., Poole P. S., Downie J. A. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet. 2011;45(1):119-144.
CrossRef - Zahran H. H. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev. 1999;63(4):968-989.
CrossRef - Shamseldin A., Abdelkhalek A., Sadowsky M. J. Recent changes to the classification of symbiotic, nitrogen-fixing, legume-associating bacteria: a review. Symbiosis 2017;71(2):91-109.
CrossRef - Christenhusz M. J., Byng J. W. The number of known plants species in the world and its annual increase. Phytotaxa 2016;261(3):201-217.
CrossRef - Bouhmouch I., Souad-Mouhsine B., Brhada F., Aurag J. Influence of host cultivars and Rhizobium species on the growth and symbiotic performance of Phaseolus vulgaris under salt stress. J Plant Physiol. 2005;162(10):1103-1113.
CrossRef - Shankar S., Haque E., Ahmed T., Kiran G. S., Hassan S., Selvin J. Rhizobia–legume symbiosis during environmental stress. In: Shrivastava N., Mahajan S., Varma A. Symbiotic Soil Microorganisms. Cham:Springer International Publishing; 2021;201-220.
CrossRef - Muresu R., Polone E., Sulas L., Baldan B., Tondello A., Delogu G., Cappuccinelli P., Alberghini S., Benhizia Y., Benhizia H., Benguedouar A., Mori B., Calamassi R., Dazzo F. B., Squartini A. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol Ecol. 2008;63(3):383-400.
CrossRef - Gehlot H. S., Panwar D., Tak N., Tak A., Sankhla I. S., Poonar N., Parihar R., Shekhawat N. S., Kumar M., Tiwari R., Ardley J., James E. K., Sprent J. I. Nodulation of legumes from the Thar desert of India and molecular characterization of their rhizobia. Plant Soil. 2012;357(1-2):227-243.
CrossRef - Sankhla I. S., Meghwal R. R., Tak N., Tak A., Gehlot H. S. Phenotypic and molecular characterization of microsymbionts associated with Crotalaria medicagenia: a native legume of the Indian Thar desert. Plant Arch. 2015;15(2):1003-1010.
- Sankhla I. S., Tak N., Meghwal R. R., Choudhary S., Tak A., Rathi S., Sprent J. I., James E. K., Gehlot H. S. Molecular characterization of nitrogen fixing microsymbionts from root nodules of Vachellia (Acacia) jacquemontii, a native legume from the Thar Desert of India. Plant Soil. 2017;410(1-2):21-40.
CrossRef - Sankhla I. S., Meghwal R. R., Choudhary S., Rathi S., Tak N., Tak A., Gehlot H. S. Molecular characterization of microsymbionts associated with root nodules of Crotalaria burhia-Ham. ex Benth., a native keystone legume species from Thar Desert of India. Indian J Exp Biol. 2018;56(6):373-384.
- Choudhary S., Meghwal R. R., Sankhla I. S., Tak N., Gehlot H. S. Molecular characterization and phylogeny of novel diverse nitrogen fixing microsymbionts associated with Vachellia (Acacia) leucophloea in arid and semi-arid regions of Rajasthan. Indian For. 2017;143(3):266-278.
- Yadav A., Solanki D., Sharma G., Dubey G., Sankhla I. S. Phenotypic and Biochemical Characterization of Rhizobia Associated with Medicago polymorpha Growing in Rajasthan. Indian Journal of Advanced Botany 2022;2(2):5-11.
CrossRef - Kasliwal S., Sankhla I. S., Mohil P. Isolation and characterization of rhizobia associated with root nodules of Butea monosperma (Lam.) taub. and Pterocarpus marsupiumJ Phytol Res. 2023;36(2):27-37.
- Singh V., Shetty B. V. A survey of natural plant resources of Rajasthan desert. Transactions of the Indian Society for Desert Technology, University Centre for Desert Studies; 1977; 2(2):296-305.
- Somasegaran P., Hoben H. J. Hand book of Rhizobia: Methods in legume-Rhizobium New York: Springer; 1994.
CrossRef - Vincent J.M. A manual for the practical study of root-nodule bacteria. Oxford: Blackwell Scientific Publications;
- Cappuccino J.G., Sherman N. Microbiology: A laboratory manual, 7th edition. California: The Benjamin/Cummings Publishing Co; 2005.
- Hungria M., Campo R., Chueire L., Grange L., Megias M. Symbiotic effectiveness of fast-growing rhizobial strains isolated from soybean nodules in Brazil. Biol Fertil Soils. 2001;33(5):387-394.
CrossRef - Kebede E., Amsalu B., Argaw A., Tamiru S. Nodulation potential and phenotypic diversity of rhizobia nodulating cowpea isolated from major growing areas of Ethiopia. Agrosyst Geosci Environ. 2022; 5(4):1-11. doi: 10.1002/agg2.20308.
CrossRef - Singh B., Kaur R., Singh K. Characterization of Rhizobium strain isolated from the roots of Trigonella foenumgraecum (fenugreek). Afr J Biotechnol. 2008;7(20):3671-3676.
- Hung M. H., Bhagwath A. A., Shen F. T., Devasya R. P., Young C. C. Indigenous rhizobia associated with native shrubby legumes in Taiwan. Pedobiologia. 2005;49(6):577-584.
CrossRef - Odori C., Ngaira J., Kinyua J., Nyaboga E. N. Morphological, genetic diversity and symbiotic functioning of rhizobia isolates nodulating cowpea (Vigna unguiculata Walp) in soils of Western Kenya and their tolerance to abiotic stress. Cognet Food Agric. 2020;6(1):1-15. doi: 10.1080/23311932.2020.1853009.
CrossRef - Wekesa C., Asudi G. O., Okoth P., Reichelt M., Muoma J. O., Furch A. C., Oelmüller R. Rhizobia contribute to salinity tolerance in Common Beans (Phaseolus vulgaris). Cells. 2022;11(22):1-18. doi: /10.3390/cells11223628.
CrossRef - Ilangumaran G., Schwinghamer T. D., Smith D. L. Rhizobacteria from root nodules of an indigenous legume enhance salinity stress tolerance in soybean. Front Sustain food syst. 2021;4:1-18. doi: 10.3389/fsufs.2020.617978.
CrossRef - Jordan, D. C. Bradyrhizobium. Bergey’s manual of systematic bacteriology. In: Krieg N. R., Holt J. G. Williams & Wilkins, Baltimore;1984:242-244.
- Abdelnaby M., Elnesairy N. N. B., Mohamed S. H., Alkhayali Y. A. A. Symbiotic and phenotypic characteristics of rhizobia nodulaing cowpea (Vigna Unguiculata Walp) grown in arid region of Libya (Fezzan). J Environ Sci Eng. 2015;4:227-239.
CrossRef - Basbuga S., Basbuga S., Yayla F., Mahmoud A. M., Can C. Diversity of rhizobial and non-rhizobial bacteria nodulating wild ancestors of grain legume crop plants. Int Microbiol. 2021;24(2):207-218.
CrossRef - Verma H., Patra R. K., Sethi D., Pattanayak S. K. Isolation and characterization of native Rhizobium from root nodules of raikia french bean growing area of Odisha. Indian J Biochem Biophys. 2022;59(9):918-926.
- Nagalingam S., Nithya T. V., Gayathri D., Sagarika A. S., Supriya G., Vidya D., Kumar B. K., Rasool A., Mir M. I. Morphological, biochemical and plant growth promoting characterization of rhizobia isolated from root nodule of Cajanus cajanPlant Arch. 2020;20(2):1293-1299.
- Oryakhil Q., Irfan A. Morphological biochemical and plant growth promoting characterization of rhizobia isolated from root nodule of Cicer arietinum. Trop Agroeco. 2020;1(2):59-63.
CrossRef - Aung T. M., Oo P. P. Isolation and characterization of Rhizobium from root nodules of Arachis hypogaea (Groundnut). J. Myanmar Acad Arts Sci. 2020;18(4B):197-210.
- Brindavathy R., Anandham R., Jamuna E., Anitha R., Hussainy S. A. H. Isolation and Screening of Efficient Rhizobial Strains and Evaluation of Their Efficiency in Moth Bean (Vigna aconitifolia Jacq). Legume Res. 2022;1-6. doi: 18805/LR-4849.
CrossRef - Pervin S., Jannat B., Sanjee S. Al. Characterization of Rhizobia from Root Nodule and Rhizosphere of Lablab purpureus and Vigna sinensis in Bangladesh. Turkish JAF Sci Tech. 2017;5(1):14-17.
CrossRef - Sadowsky M. J., Keyser H. H., Bohlool B. B. Biochemical characterization of fast-and slow-growing rhizobia that nodulate soybeans. Int J Syst Evol Microbiol. 1983;33(4):716-722.
CrossRef - Negash D., Tulu D., Alazar E., Chris O. Phenotypic characteristics of rhizobial and non-rhizobial isolates recovered from root nodules of chickpea (Cicer arietinum) grown in Ethiopia. Afr J Microbiol Res. 2018;12(4):73-85.
CrossRef - Wanore D. S., Bachore M. A., Eromo E. K., Nure R. K., Bidiko G. B., Abdo S. S. Isolation and Characterization of Nitrogen Fixing Bacteria from Rhizospher of Chickpea and Pea nodules, Hossana, Ethiopia. Uttar Pradesh Journal of Zoology. 2023;44(15):97-107.
CrossRef - Hamza T. A., Alebejo A. L. Isolation and characterization of rhizobia from rhizospher and root nodule of cowpea, elephant and lab lab plants. Int J Nov Res Interdiscip Stud. 2017;4(4):1-7.
- Rajpoot P., Panwar K. S. Isolation & characterization of rhizobia and their effect on Vigna radiataOcta J Biosci. 2013;1(1):69-76.
- Purwaningsih S., Agustiyani D., Antonius S. Diversity, activity, and effectiveness of Rhizobium bacteria as plant growth promoting rhizobacteria (PGPR) isolated from Dieng, Central Java. Iran J Microbiol. 2021;13(1):130-136.
CrossRef - Rathi S., Gaur S., Tak N., Tak A., Gehlot H. S. Genetically diverse root nodule bacteria associated with Alysicarpus vaginalis from alkaline soil of Rajasthan, India. Plant Arch. 2017;17(1):495-505.
- Abera Y., Masso C., Assefa F. Phenotypic, Host Range and Symbiotic. Ethiop J Agric Sci. 2018;28(3):95-116.
- Beshah A., Assefa F., Muleta D., Legese G. Screening groundnut rhizobia for multiple plant growth promoting activities in Ethiopia. Afr J Microbiol Res. 2023;17(9):212-220.

